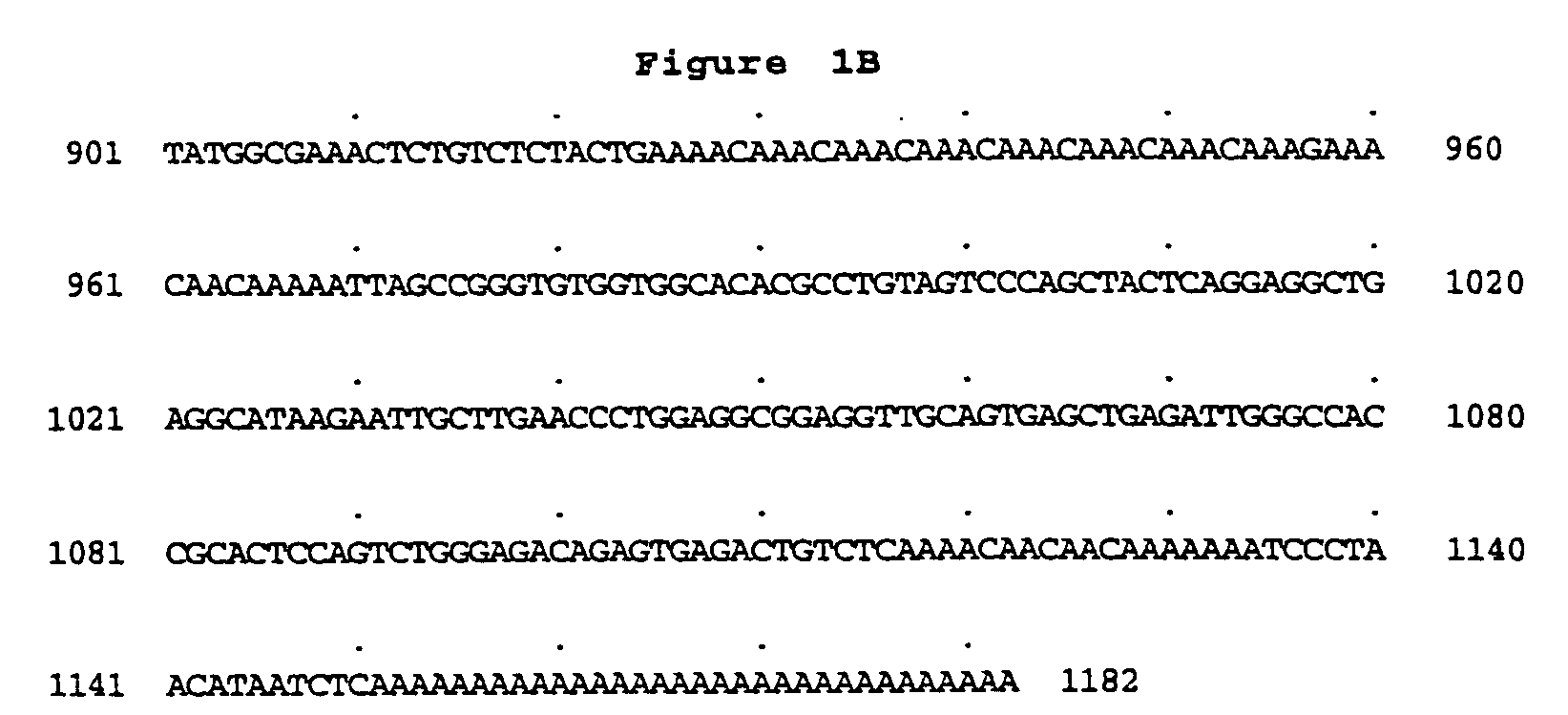
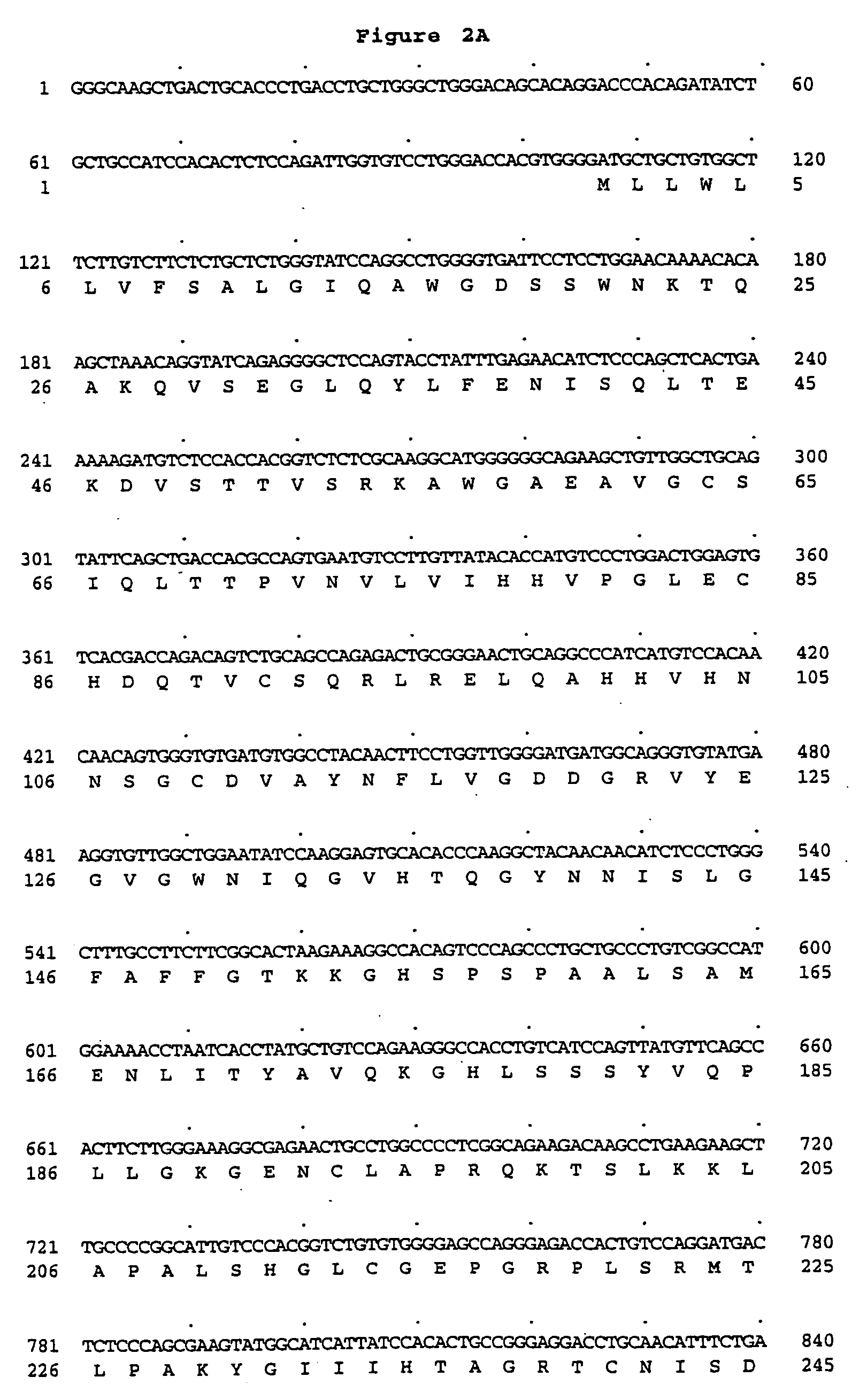
Peptidoglycan recognition proteins
a technology of peptidoglycans and recognition proteins, applied in the field of new drugs, can solve the problems of weight loss in cancer patients, and achieve the effect of regulating the growth activity of keratinocytes and preventing septic shock
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Isolation of PGRP-K, PGRP-W, and / or PGRP-C cDNA Clone(s) from the Deposited Sample(s)
[0303] The cDNA for PGRP-K (ATCC Accession No: 203564) is inserted into the Sal I and Not I multiple cloning site of pCMVSport 2.0 (Life Technologies). pCMVSport 2.0 contains an ampicillin resistance gene and may be transformed into E. coli strain DH10B, available from Life Technologies. (See, for instance, Gruber, C. E., et al., Focus 15:59-(1993).)
[0304] The cDNA for PGRP-W (ATCC Accession No: 203563) is inserted into the Sal I and Not I multiple cloning site of pCMVSport 3.0 (Life Technologies). pCMVSport 3.0 contains an ampicillin resistance gene and may be transformed into E. coli strain DH10B, available from Life Technologies. (See, for instance, Gruber, C. E., et al., Focus 15:59-(1993).)
[0305] The cDNA for PGRP-C (ATCC Accession No: 209683) is inserted into the EcoRI and Xho I multiple cloning site of Uni-Zap XR (Stratagene). Uni-Zap XR contains an ampicillin resistance gene and may be tr...
example 2
Isolation of PGRP-K, PGRP-W, or PGRP-C Genomic Clones
[0313] A human genomic P1 library (Genomic Systems, Inc.) is screened by PCR using primers selected for the cDNA sequences corresponding to SEQ ID NO:1, SEQ ID NO:3, or SEQ ID NO:5, according to the method described in Example 1. (See also, Sambrook.)
example 3
Chromosomal Mapping of PGRP-K, PGRP-W, or PGRP-C
[0314] An oligonucleotide primer set is designed according to the sequence at the 5′end of SEQ ID NO:1, SEQ ID NO:3, or SEQ ID NO:5. This primer preferably spans about 100 nucleotides. This primer set is then used in a polymerase chain reaction under the following set of conditions: 30 seconds, 95 degree C.; 1 minute, 56 degree C; 1 minute, 70 degree C. This cycle is repeated 32 times followed by one 5 minute cycle at 70 degree C. Human, mouse, and hamster DNA is used as template in addition to a somatic cell hybrid panel containing individual chromosomes or chromosome fragments (Bios, Inc). The reactions is analyzed on either 8% polyacrylamide gels or 3.5% agarose gels. Chromosome mapping is determined by the presence of an approximately 100 bp PCR fragment in the particular somatic cell hybrid.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com