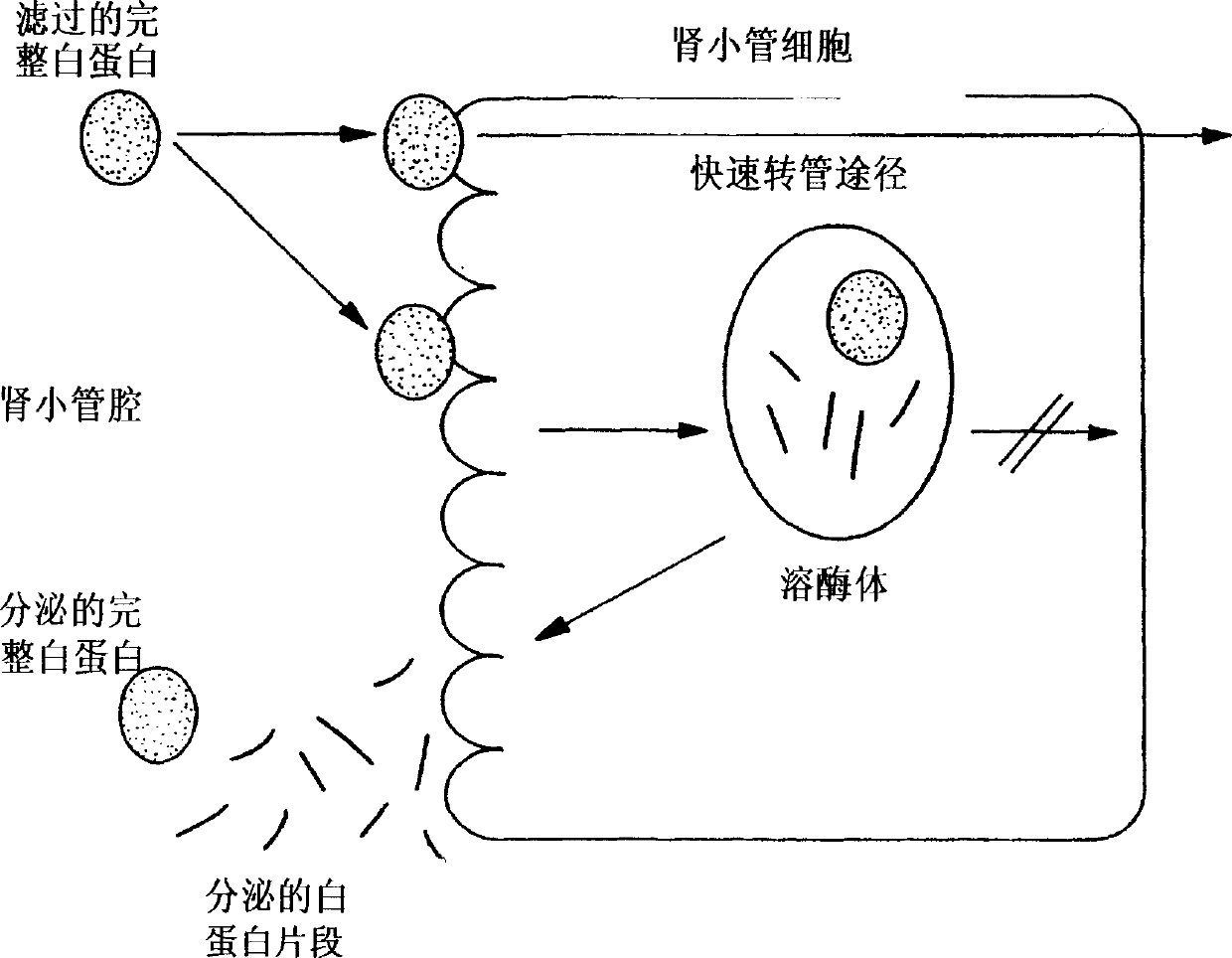
Method for kidney disease detection by protein profiling
A protein map and kidney disease technology, applied in the field of kidney disease complications, can solve problems such as inability to detect quickly, disappointing effectiveness, and unrepeatable technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0139] Example 1: Size Exclusion Chromatography of Human Serum Albumin (HSA)
[0140] The distribution of albumin in urine was analyzed using urine samples provided by normal healthy volunteers.
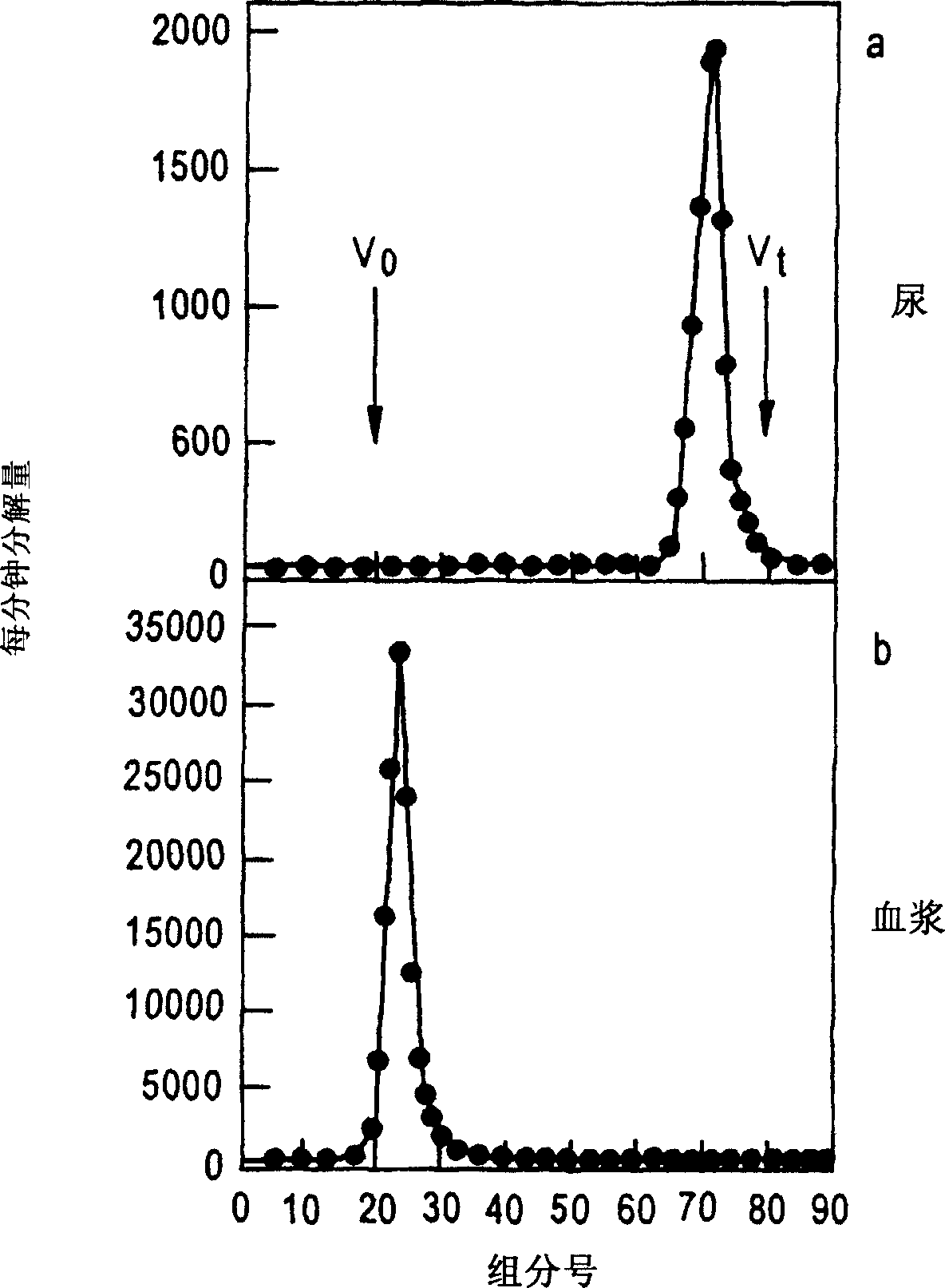
[0141] Will 3 H[HAS] (human serum albumin) was injected into healthy volunteers, urine samples and plasma were collected, and they were analyzed by size exclusion chromatography with G-100 column. The column was eluted with PBS (pH=7.4) at 4°C at a rate of 20 mL / hr. The external water volume (Vo) of the column was determined with blue dextran T2000, while the total volume was determined with tritiated water.
[0142] Tritium radioactivity in 1 mL of aqueous samples was determined with 3 mL of scintillator and detected on a Wallac 1410 liquid scintillation counter (Wallal Turku, Finland).
[0143] Figure 2 shows the distribution of albumin in urine and plasma.
Embodiment 2
[0144] Example 2: Albumin secretion in normal healthy volunteers and diabetic patients
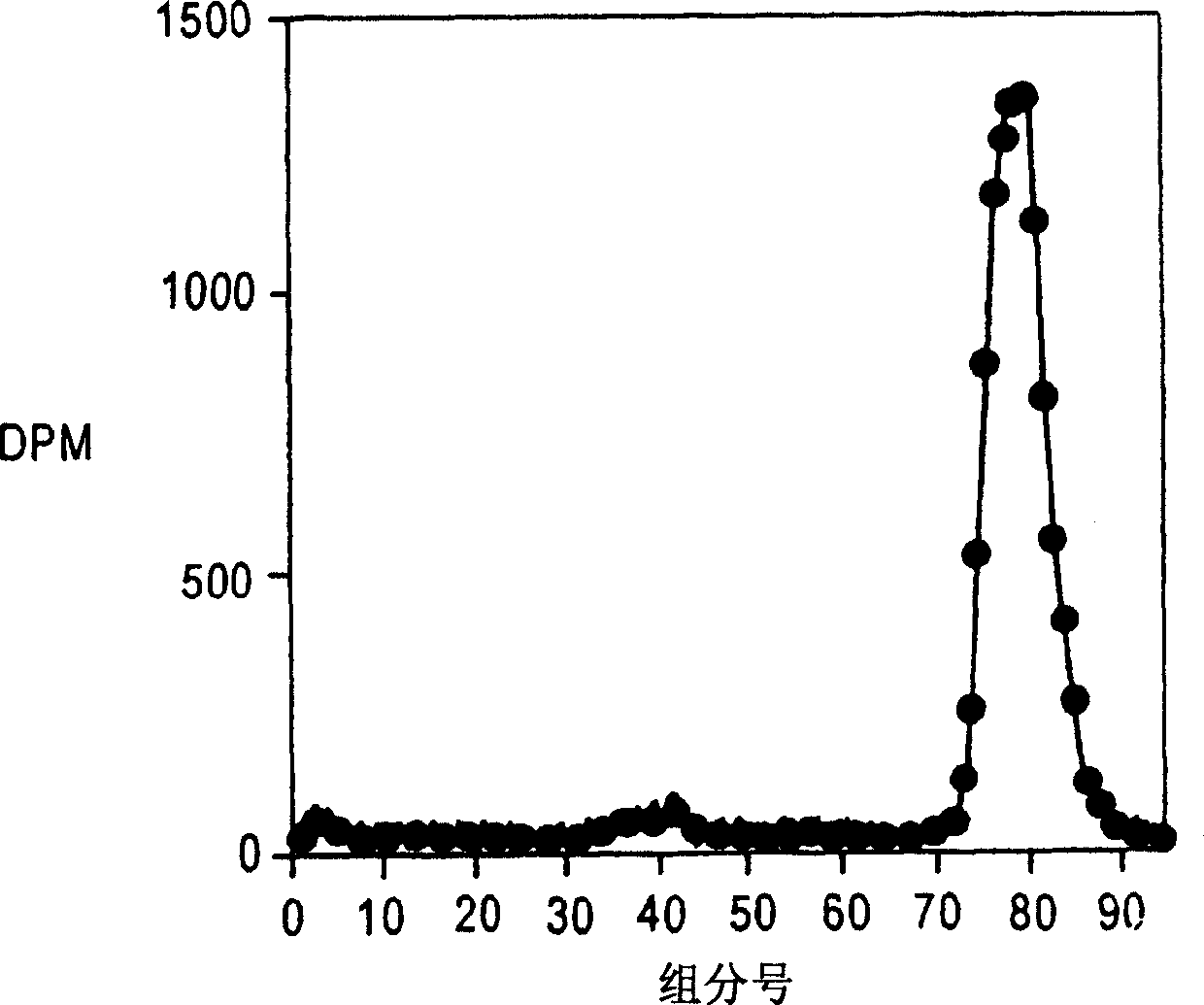
[0145] Will use in embodiment 1 3 H[HAS] was injected into normal healthy volunteers and diabetic patients. Collect urine sample, determine with the method of embodiment 1 3 H[HAS].
[0146] Normal healthy volunteers ( FIG. 3 ) were shown to secrete fragmented albumin on size exclusion chromatography performed as in Example 1 .
[0147] Diabetic patients (Figure 4) showed the presence of essentially full-length and fragmented albumin on size exclusion chromatography. However, albumin secretion rates detected with these methods were of the order of 5 μg / min (control) and 1457 μg / min (diabetic patients).
Embodiment 3
[0148] Example 3: Determination of intact albumin and intact / modified albumin on HPLC
[0149] Urine samples were collected from normal healthy volunteers, normoalbuminuric diabetics and macroalbuminuric patients. Collect midstream urine into a 50 μL urine sample container. Urine samples were frozen until further use. Urine samples were centrifuged at 5000g prior to HPLC analysis.
[0150] Urine samples were analyzed by HPLC with a hydrophobic column Zorbax 300SB-CB (4.6mm×150mm). Utilize a 50 µL sample loop.
[0151] The sample was eluted from the column using the following conditions
[0152] Solvent A Water 1% trifluoroacetic acid
[0153] Solvent B 60% acetonitrile, 0.09% TFA
[0154]Solvent A2 99.96 > 00.00: 49.58 minutes
[0155] Pressure 9.014M Pascal (~1100psi).
[0156] Solvent B2 0.04>100.0: 49.58 minutes
[0157] Pressure 7.154M Pascal
[0158] The wavelength used is 214 nm.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com