Novel deoxyribonucleside kinase enzyme multi-substrate variants
A deoxyribonucleoside kinase, enzyme variant technology, applied in the direction of enzymes, microorganisms, transferases, etc., can solve problems such as major by-products
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0254] PCR-induced Dm-dNK variants
[0255] A directed evolution approach based on mutagenesis PCR was used to generate mutant kinase forms. The open reading frame (ORF) of Dm-dNK was mutagenized by different nucleotide analog concentrations and the effect of different nucleotide analog concentrations was studied. The mutagenized PCR fragment was ligated into an expression plasmid and then transformed into a TK-deficient E. coli KY895 strain.
[0256] Random Mutagenesis and Mutation Library Construction
[0257] Expression vector pGEX-2T-rDm-dNK [Munch-Petersen et al. J. Biol. Chem. 2000 275(9) 6673-6679] was used as template for PCR mutagenesis.
[0258] The open reading frame (ORF) of Dm-dNK was amplified using the following primers:
[0259] Dm-TK3: 5'-CGCGGATCCATGGCGGAGGCAGCATCCT-3' (SEQ ID NO: 7); and
[0260] Dm-TK4: 5'-CGGAATTCTTATCTGGCGACCCTCTGGCGT-3' (SEQ ID NO: 8).
[0261] PCR is performed in two steps. PCR was first performed in a 20 μl reaction with 0.15...
Embodiment 2
[0273] Characterization of enzyme variants
[0274] sequencing
[0275] Using Thermo Sequenase radio-labelled terminator cycle sequencing kit and P 33 Purified plasmids were sequenced manually with tagged ddNTPs (Amersham Corp.) by the Sanger dideoxy method.
[0276] Determination of LD 100 (in vivo characterization)
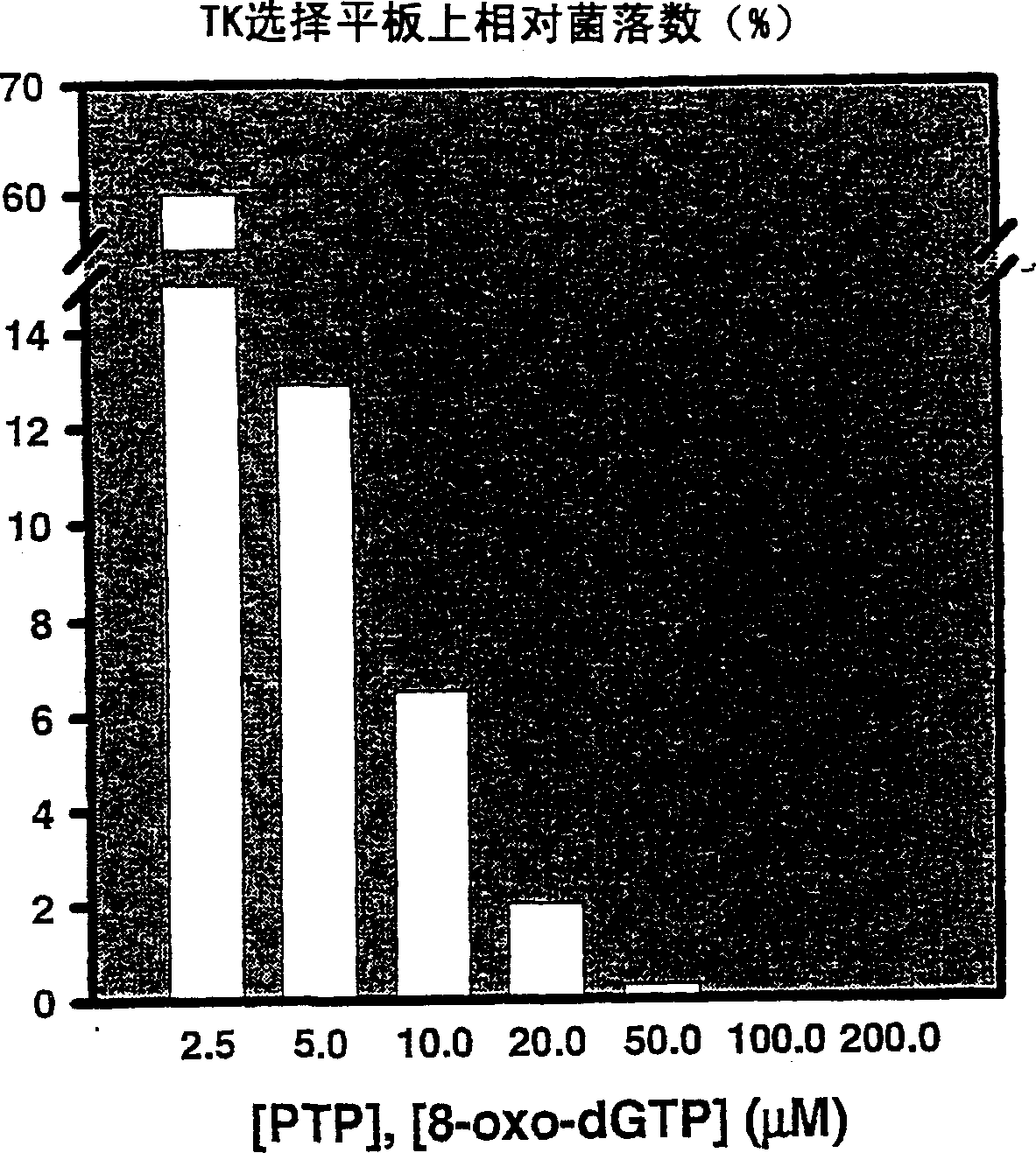
[0277] To determine the lethal dose (LD) of nucleoside analogues 100 ) (at this lethal dose, the bacteria did not grow), logarithmic dilutions of the nucleoside analogs were performed and all clones showing increased sensitivity to at least one nucleoside analog were assayed on M9 plates. Plates with concentrations ranging from 10-1000 μM AraA, 3.16-1000 μM AraC; 0.01-100 μM AZT; 0.316-31.6 μM ddA; 0.0316-100 μM 2CdA or 10-3500 μM ddC were used to determine the LD of putative mutants 100(concentration capable of causing 100% death).
[0278] Plates were prepared by mixing media and the like at the lowest temperature possible prior to pouring the plat...
Embodiment 3
[0292] sequence determination
[0293] Publicly available expressed sequence tag (EST) libraries were searched with the basic local alignment search tool (BLAST) in the GenBank database of the National Institute for Biotechnology information to identify ESTs encoding enzymes similar to Dm-dNK (GenBanK ACCN AF226281) . Through this method, the ESTs ACCN AU004911 from Bombyx mori and ACCN AW159435 from Xenopus laevis were identified.
[0294] ESTs were obtained from Genome Research Group, National Institute of Radiological Sciences, Anagawa 4-9-1, Inage, Chiba 263-8555, Japan (ACCN AU004911) and Lita Annenberg Hazen Genome Sequencing Center, Cold Spring Harbor Laboratory, PO Box 100 , Cold Spring Harbor, NY 11724, USA (AW159435). The complete open reading frames of the deoxyribonucleoside kinases encoded by these two ESTs were determined by DNA sequencing (see Example 2).
[0295] The complete open reading frame was then submitted to GenBank and assigned numbers ACCNAF226281 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com