High temp. resistant guanylate kinase gene, its coded polypeptide and preparing method thereof
A high temperature resistant, guanylic acid technology, applied in the field of mutation or genetic engineering, can solve problems affecting binding objects, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
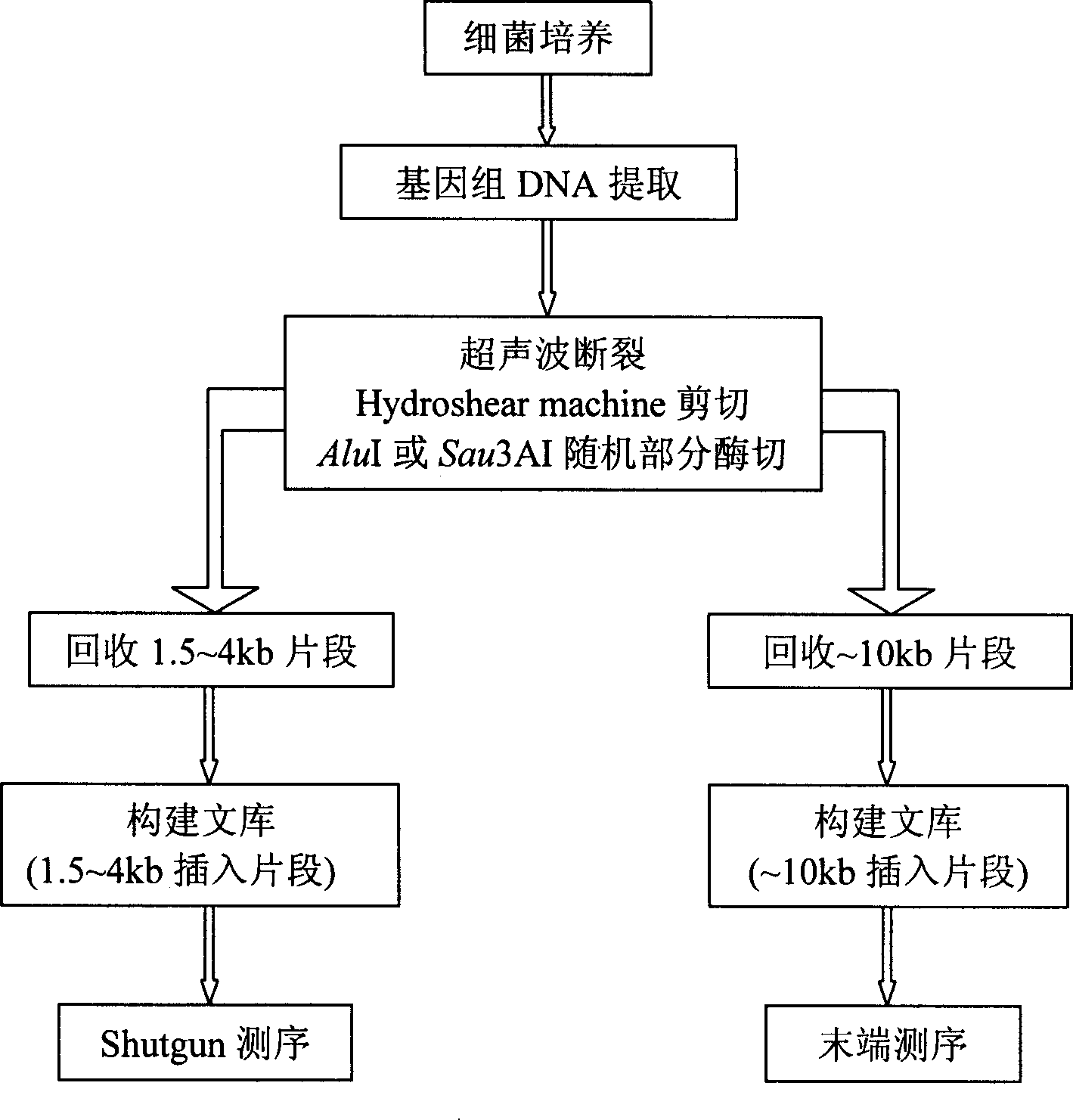
[0038] Example 1: Construction of a sequencing library
[0039] The sequencing library was constructed using the genome-wide shotgun method. Firstly, Tengchong thermophilic anaerobic bacteria were cultured according to (Yanfen Xue, 2000) modified MB medium (Balch et al., 1979), bacteria were collected according to Marmur (1961) method, and total DNA was extracted. In order to ensure the randomness of sequencing library construction and avoid the problem of breaking hot spots to the greatest extent, the Dolly method and the principle of library construction under different conditions were adopted. First, physical shearing methods (including ultrasonic method and HydroshearMachine shearing) were used, and then AluI was selected for random partial enzyme digestion according to the genome characteristics of the bacteria. Different intensities are used to treat rods during physical shearing, and samples are treated by setting enzyme gradients during enzyme digestion. After the pr...
Embodiment 2
[0040] Example 2: Genome Sequencing
[0041] When sequencing the genome of thermophilic anaerobic bacteria in Tengchong, two automatic sequencers were mainly used: ABI377 and MegaBACE 1000. Both sequencers use the principle of electrophoresis for sequencing (see figure 2 ), 96 samples can be completed each time. ABI377 is a product of PE Company, which is a kind of ABI series. It belongs to the slab gel electrophoresis sequencer. MegaBACE 1000 is a product of Pharmacia, which belongs to capillary gel electrophoresis sequencer.
Embodiment 3
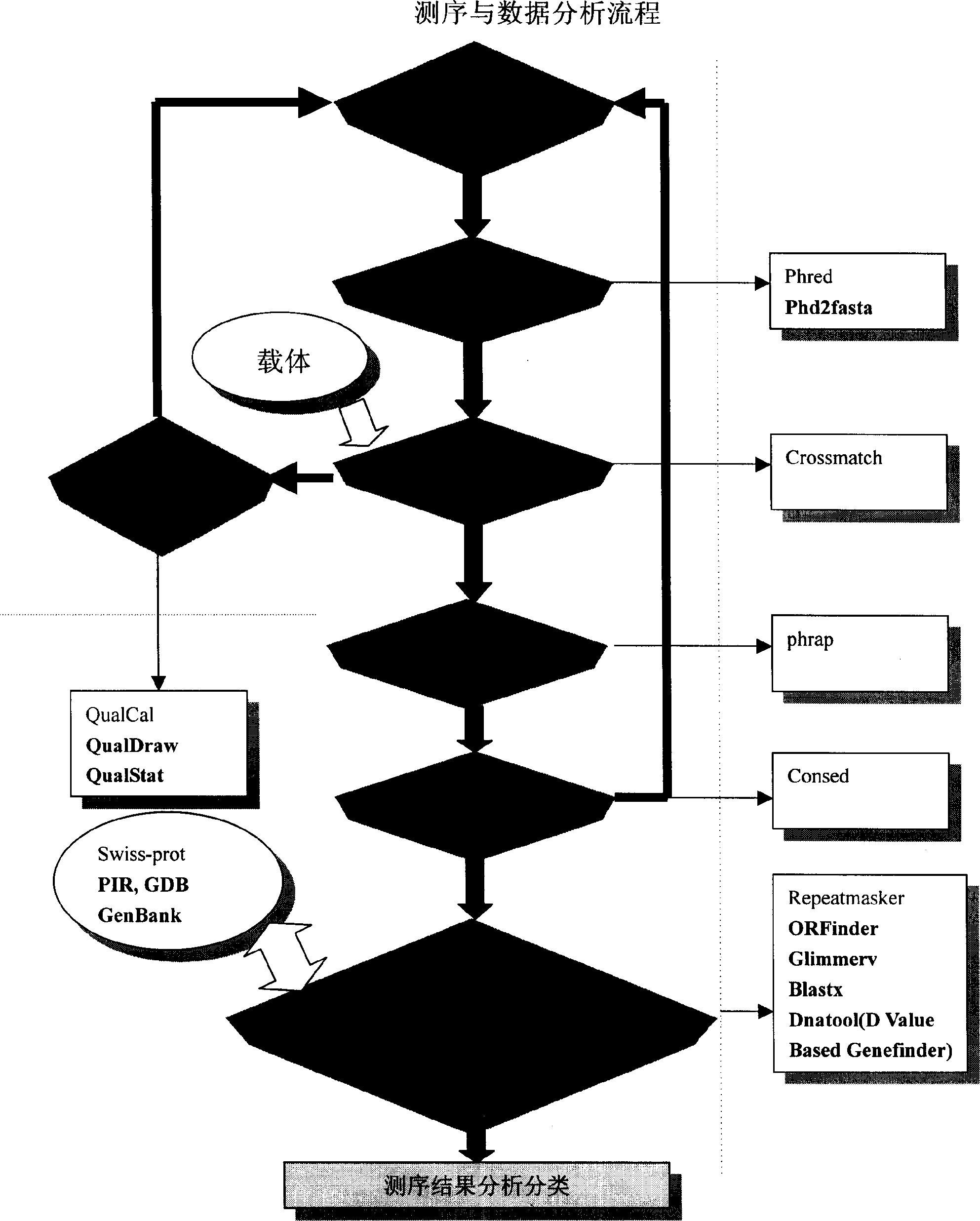
[0042] Example 3: Basecalling and sequencing quality monitoring
[0043] The so-called basecalling refers to the process of obtaining the correct base sequence from the raw data file obtained on the sequencer. Since the sequencer obtains the intensity change traces (traces) of light of different wavelengths corresponding to the four bases A, T, G, and C, it is necessary to use a computer to adopt a certain algorithm to correctly identify the bases corresponding to the different traces. . We used Phred software (Ewing B, Hillier L, 1998) because its results are more reliable and its output is more convenient for further analysis by other programs in the same software package.
[0044] The algorithm principle of Phred's basecalling is to judge the base type based on the shape, distance, and signal-to-noise ratio of each peak in the trajectory, and at the same time give the credibility information for this base, that is, the sequencing quality of the base. In large-scale sequen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com