Multi-nucleic acid aptamers for detecting pathogenic microorganisms and combinations thereof
A technology of pathogenic microorganisms and nucleic acid aptamers, applied in measuring devices, recombinant DNA technology, instruments, etc., can solve problems such as evaluating the ability of nucleic acid aptamers to bind to other proteins, and achieve convenient modification and substitution, small molecular weight, The effect of good affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Preparation of nucleic acid aptamers
[0061] 1. Construction of screening library
[0062] The ssDNA library used in this study is 76 nt in full length, consisting of 20 nt fixed primer fragments at both ends and a 30 nt random fragment in the middle; the 5' FITC fluorophore of the forward primer is modified with the 5' of the original library. The fixed sequence at the ' end is the same; the 5' end of the reverse primer is labeled with Biotin, and the sequence is reverse complementary to the fixed sequence at the 3' end of the original library;
[0063]The sequence of the screening library targeting SARS-CoV-2 nucleocapsid protein is:
[0064] SEQ ID NO. 5:
[0065] CTTCTGCACGCCTCCTTCC (N 35 ) GGAGACGAGATCGGCGGACACT.
[0066] The primers used to construct the library are:
[0067] Forward primer: SEQ ID NO.6: CTTCTGCACGCCTCCTTCC;
[0068] Reverse primer: SEQ ID NO. 7: AGTGTCCGCCGATCTCGTCTCC.
[0069] 2. Conjugation of protein and magnetic beads
[0070] Take s...
Embodiment 2
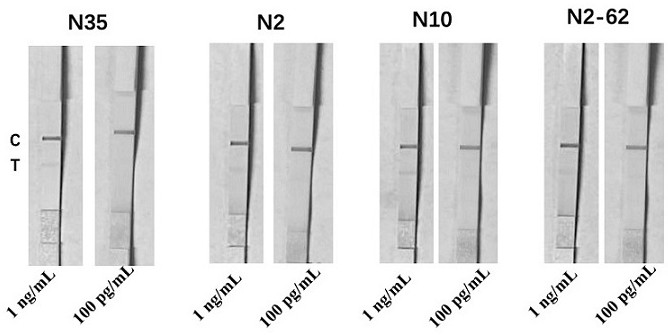
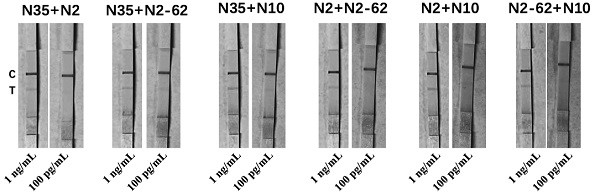
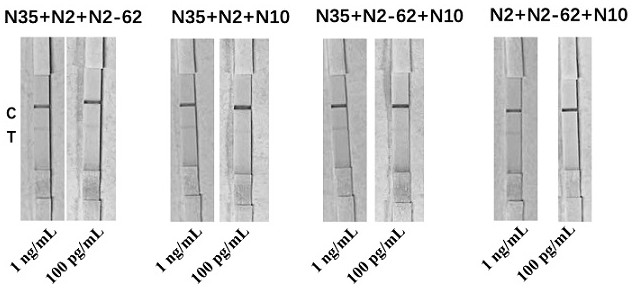
[0087] Sandwich combination selection of different aptamers and antibodies
[0088] After testing and screening, the selected nucleic acid aptamer combinations are shown in Table 1.
[0089] Table 1 Nucleic acid aptamer combination scheme
[0090] group combination 1 N35 / N2 2 N35 / N2-62 3 N35 / N10 4 N2 / N2-62 5 N2 / N10 6 N2-62 / N10 7 N35 / N2 / N2-62 8 N35 / N2 / N10 9 N35 / N2-62 / N10 10 N2 / N2-62 / N10 11 N35 / N2 / N10 / N2-62
Embodiment 3
[0092] Preparation of Nucleic Acid Aptamers Labeled with Latex Microspheres
[0093] The streptavidin (SA) modified latex particles (mass fraction 1%, provided by Hangzhou Ustar) were diluted twice with ultrapure water to a mass fraction of 0.5%. Then, a 4-fold excess of biotin-modified nucleic acid aptamer (biotin-modified nucleic acid aptamer was synthesized by Shanghai Sangon Biotechnology Co., Ltd.) was added to the SA-modified latex particles with a mass fraction of 0.5% for cross-linking. The obtained cross-linked product was centrifuged to remove nucleic acid aptamers unbound to latex particles in the supernatant, and the centrifugation conditions were 4°C, 10000 rpm, 10 min. Then, the cross-linked product was washed twice with ultrapure water by centrifugation, and the final volume was made up with 50 μL of dispensing solution for subsequent use.
[0094] Among them, there are two marking methods, namely:
[0095] 1) Mixed labeling, that is, different nucleic acid ap...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com