Highland barley drought-resistant related gene and kit and method for identifying drought-resistant highland barley
A kit and highland barley technology, applied in botany equipment and methods, biochemical equipment and methods, genetic engineering, etc., to achieve the effect of simple method, low sequencing length and low requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1, identification of drought-resistant highland barley
[0027] First, synthesize primer pairs for drought resistance genes:
[0028] F: CACGGGATGCGACAGAGACC (SEQ ID NO. 2);
[0029] R: GCTGATCCGGCAATCCGAGC (SEQ ID NO. 3).
[0030] Use the following methods to identify:
[0031] a) Using the TGuide OSR-M301 kit of Tiangen Biochemical Technology (Beijing) Co., Ltd. to extract the total genomic DNA of the highland barley leaf tissue to be inspected;
[0032] b) using the total genomic DNA described in step a) as a template, using the synthesized primer pair to carry out a PCR reaction, wherein the reaction system is 25 μL, and the annealing temperature is 60°C;
[0033] c) Perform 1% agarose gel electrophoresis detection on the PCR product obtained in step b), and observe a bright band of about 3Kb;
[0034] d) Sanger sequencing by capillary electrophoresis is performed on the PCR product of step c) to obtain a gene sequencing peak map of SEQ ID NO: 1;
[...
Embodiment 2
[0037] Example 2 Genetic research on drought resistance of highland barley
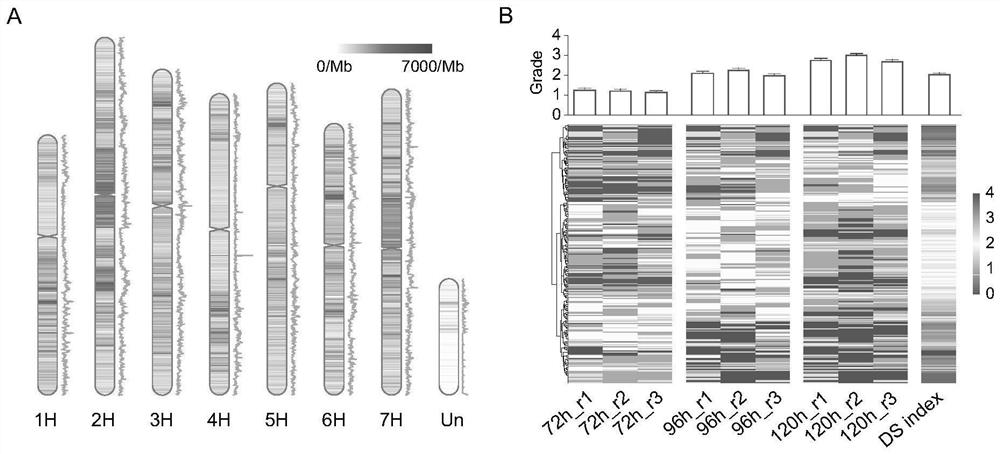
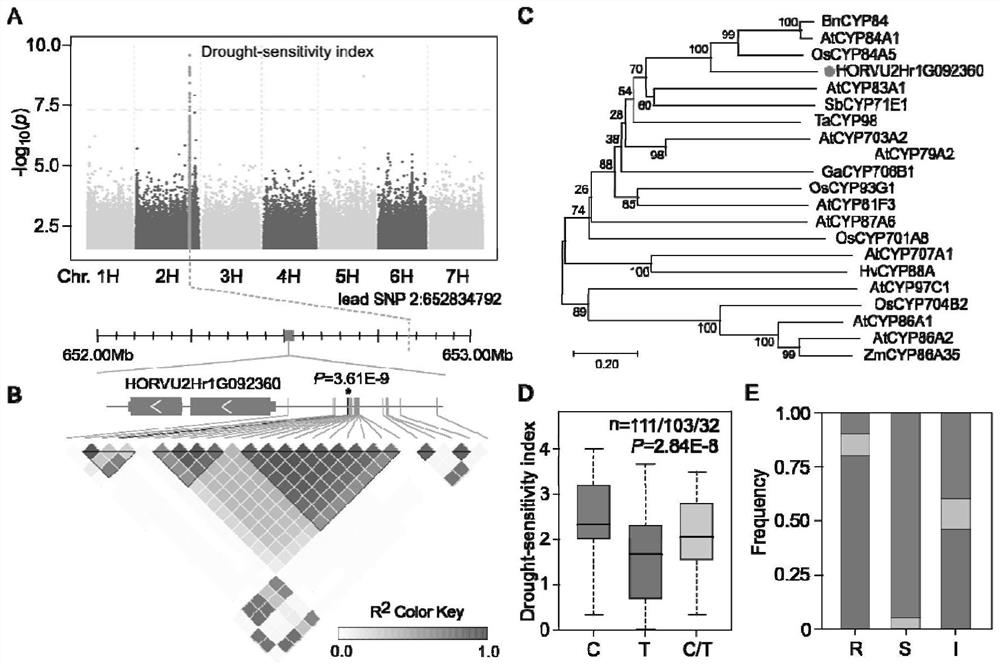
[0038] The present invention collects 246 highland barley germplasm resources from all over the world, extracts DNA and obtains resequencing data based on second-generation sequencing. Based on the Illumina Hiseq 2000 sequencing platform, a total of 10.44Tb of sequencing data were obtained, and the coverage rate of each sample exceeded 8.5. These reads were mapped to the IBSC_v2 reference genome (Mascher et al., 2017), which was well assembled and annotated, with an average read mapping rate of approximately 86.69% across all samples. After SNP Calling, the present invention selected SNPs with deletion calling frequency less than 10% and minor allele frequency greater than 5%, resulting in a total of 7,744,571 SNPs ranging from 893,125 (chr1H) to 1,508,037 (chr2H) per chromosome, each The maximum number of SNPs for Mb is 6,979. In addition, the nucleotide diversity π (window 1Mb) ranged from 2.2E-07...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com