Visual violet light cycle simulation method based on molecular dynamics
A molecular dynamics and rhodopsin technology, applied in the field of neuroscience, can solve problems such as insufficient sampling and inability to accurately obtain ion channels, and achieve the effect of improving insufficient sampling
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0040] The technical solutions in the embodiments of the present invention will be described clearly and in detail below with reference to the accompanying drawings in the embodiments of the present invention. The described embodiments are only some of the embodiments of the invention.
[0041] The technical scheme that the present invention solves the above-mentioned technical problems is:
[0042] To achieve the above object, the present invention adopts the following technical scheme: a molecular dynamics-based rhodopsin light cycle simulation system, comprising the following steps:
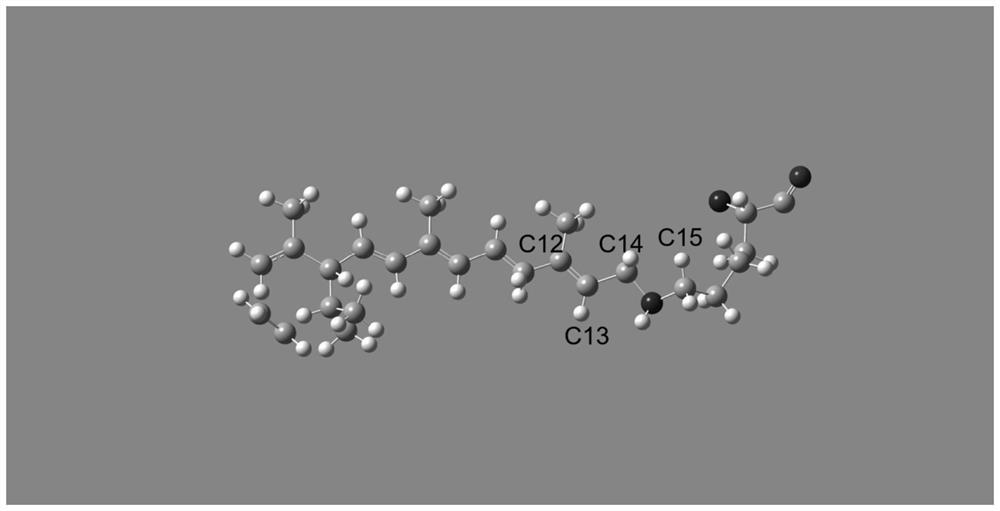
[0043] Step 1. Use Gaussian software to twist and adjust the dihedral angle of the chromophore C12-C13=C14-C15; the protein multimer file downloaded from the PDB database is manually deleted to turn into a monomer crystal , retain single-chain amino acid residues, delete all other small molecules, and other structures. Load the monomer PDB file using Gaussian software. The structure of chro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com