Sequence matching method based on k-mer
A sequence and base technology, applied in the field of sequence alignment based on k-mer, can solve the problems of large memory occupation and slow processing speed, so as to meet the alignment results, improve efficiency and accuracy, and save computing resources and time. cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0047] To better illustrate the present invention, the following in conjunction with the embodiments to be further described, examples of the embodiments are shown in the accompanying drawings. The embodiments described below by reference to the accompanying drawings are exemplary and are intended to be used to explain the present invention, and cannot be construed as a limitation of the present invention.
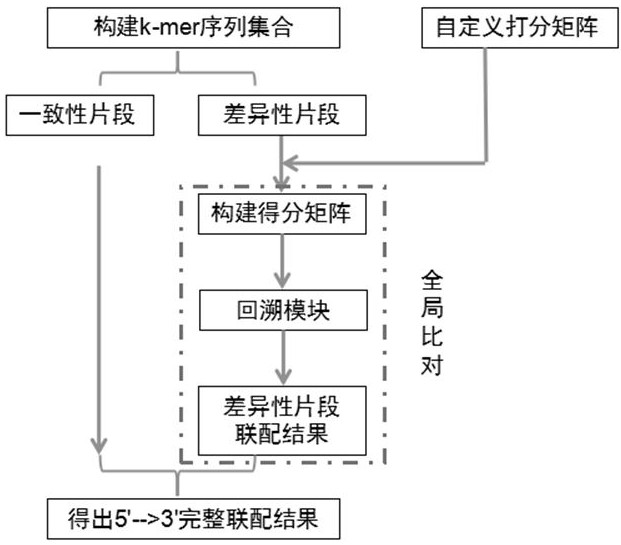
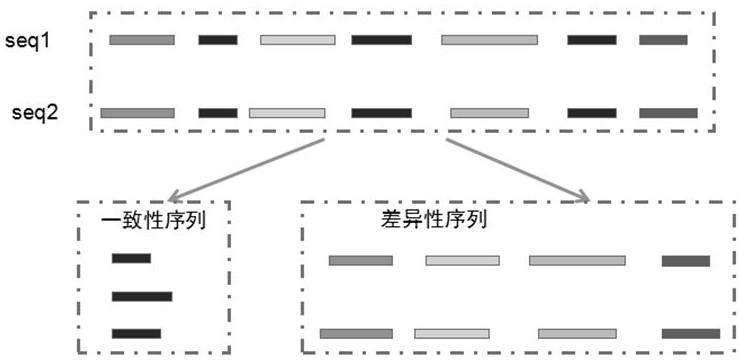
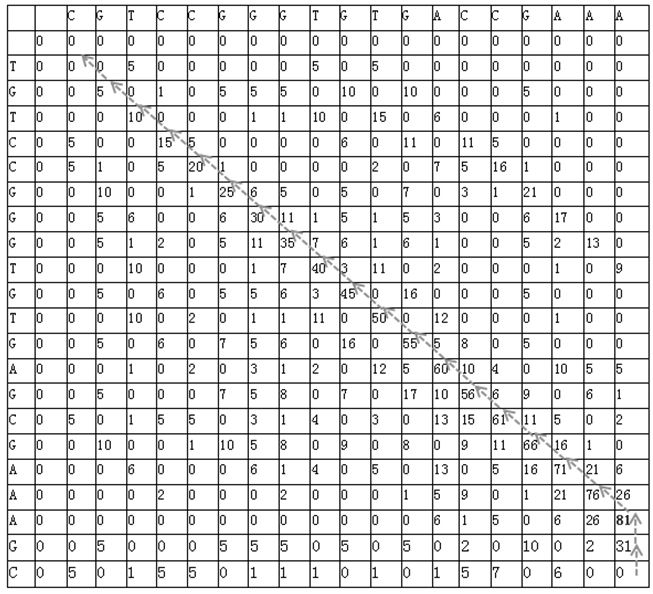
[0048] The sequence combination method provided by the present invention is based on the core idea of global comparison, on the basis of which the k-mer idea is further introduced, in accordance with custom scoring and specific backtracking rules, to achieve the combination of sequences. as Figure 1 As shown, the specific core steps are as follows: the first step, respectively, the input seq1 sequence and seq2 sequence according to the first predetermined length k -mer analysis, obtained k -mer sequence set, described k -mer is predeterminedly longer than the number of mismatc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com