A dual-aptamer functional nucleic acid thermostatic microfluidic chip sensor for microbial detection
A microfluidic chip and functional nucleic acid technology, which is applied in the fields of biological material analysis, laboratory containers and instruments, etc., can solve the problems of time-consuming, inability to detect on-site, cumbersome operation, etc., to improve the capture ability and efficient on-site detection. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0089] Example 1 Microfluidic chip biosensor composition, installation instructions and use of each part
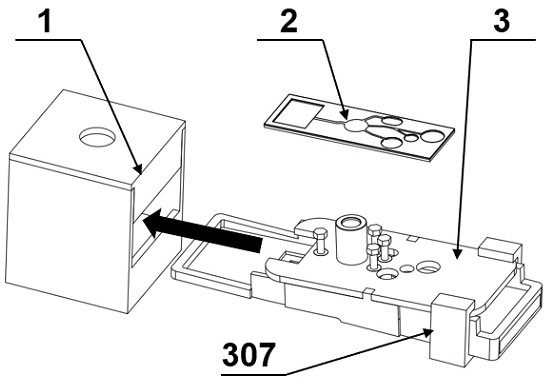
[0090] The microfluidic chip sensor is composed of figure 1 As shown, it includes an observation dark box 1 for observing fluorescence, a microfluidic chip 2 for detecting Bacillus cereus, and a capillary microvalve control device 3 for controlling the flow of liquid in the microfluidic chip. Insert the microfluidic chip capillary valve control device 3 with the microfluidic chip 2 installed into the fluorescence collection dark box 1 .
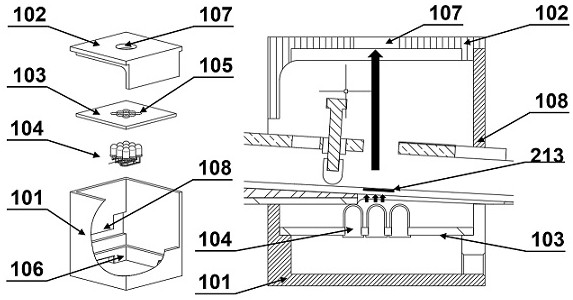
[0091] Observation obscura 1 for observing fluorescence such as figure 2 As shown, it is composed of a box body 101 , a dark box top cover 102 , a lamp bead partition 103 and a lamp bead 104 . The lamp beads 104 are installed in the 9×9 array holes 105 reserved on the lamp bead separator 103 , and welded to form a lamp bead array 104 that generates sufficient fluorescent intensity. Install the lamp bead spacer 103 on which the lamp b...
Embodiment 2
[0095] Example 2 Screening of recognition elements in microfluidic chip biosensors
[0096] The nucleic acid aptamer of Bacillus cereus is obtained by whole-cell screening technology, thereby realizing the specific recognition of Bacillus cereus in the present application. The nucleic acid sequences used in the experiment are shown in Table 1. The specific whole-cell screening process is as follows: (a) Take 1 nmol of the synthesized random library, denature it at 95°C for 5 minutes, immediately place it on ice, and place it for 5 minutes. Add 600 μL of bacterial suspension and excess tRNA and BSA to the treated library. Incubate at 35°C with shaking at 120rpm for 90min. (b) After incubation, centrifuge at 4°C for 5 min at 8000 rpm. The ssDNA bound to the target cell is settled down with the cell, while the ssDNA that is not bound to the target cell is in the supernatant. Wash the pellet twice with 1×BB to remove ssDNA with weak binding ability. (c) Add 100 μL of 1×TE buffe...
Embodiment 3
[0102] Example 3 Cutting optimization of identification elements in microfluidic chip biosensors
[0103] On the basis of the results of Example 2, cutting optimization was carried out, using IDT to predict the secondary structure of the preferred aptamer, selecting the predicted result with the lowest minimum free energy for subsequent cutting operations, and reserving 2-3 nt on both sides of the hairpin structure. At the end, the trimmed aptamer is called a trimmed aptamer. Then, the cutting aptamer is cut again. The purpose of the second cutting is to disassemble and subdivide the complete stem-loop structure in the cutting aptamer, so as to accurately locate the core sequence of the aptamer participating in the interaction, and A secondary cropped aptamer is called a secondary cropped aptamer. The IDT website was used to predict the secondary structure of the tailored aptamer and the secondary tailored aptamer, and the sequence with suitable cutting length and lower minim...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com