Application of non-conformable lentiviral vector system in gene editor delivery
A lentiviral vector, non-integrated technology, applied in applications, genetic engineering, plant genetic improvement, etc., can solve problems such as limited packaging capacity, insertion mutation, and inability to accommodate base editors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
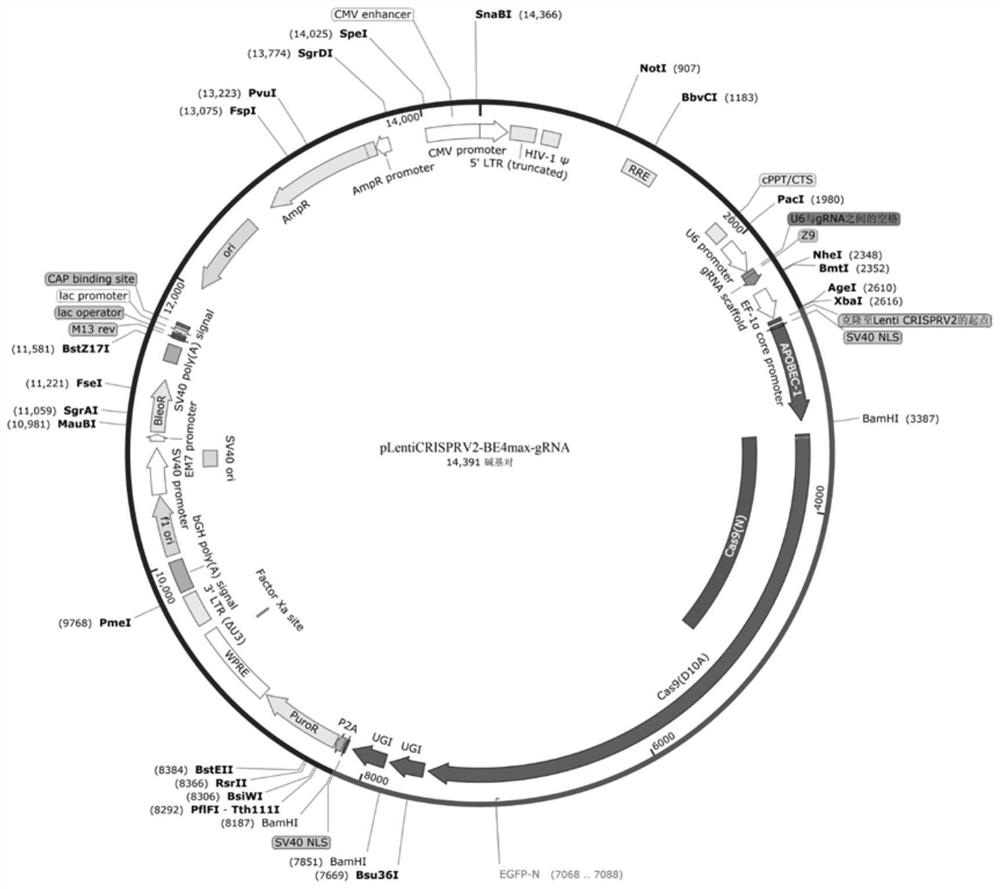
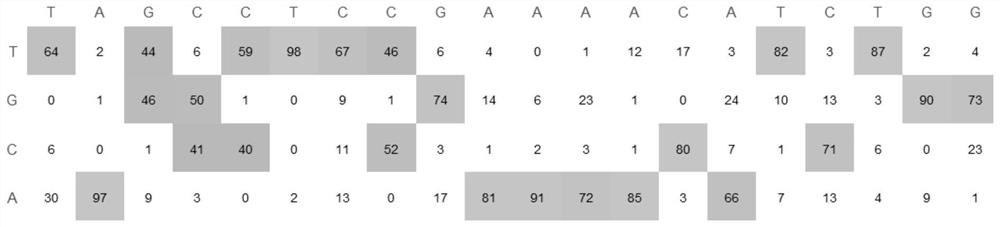
[0067] Example 1. Non-integrating lentivirus delivery CBE realizes editing of specific sites
[0068] It is a relatively safe and efficient method to use non-integrating lentiviral packaging CBE base editors to infect target cells after harvesting virus particles to achieve gene editing at specific sites.
[0069] Experimental process: The coding genes of the currently commonly used BE4max and hyeA3ABE3max CBE base editors were respectively constructed into lentiviral vectors, and the puromycin gene was expressed in the vectors, which can be used for subsequent screening. In addition, the corresponding gRNA sequence was inserted behind the N-terminal U6 promoter through the Golden gate.
[0070] The specific construction method is as follows:
[0071] Step 1: Use the full-length plasmid of the BE4max base editor expression vector as a template, and use the following primer pair BE4max-F / BE4max-R as amplification primers to perform PCR amplification to obtain the target fragme...
Embodiment 2
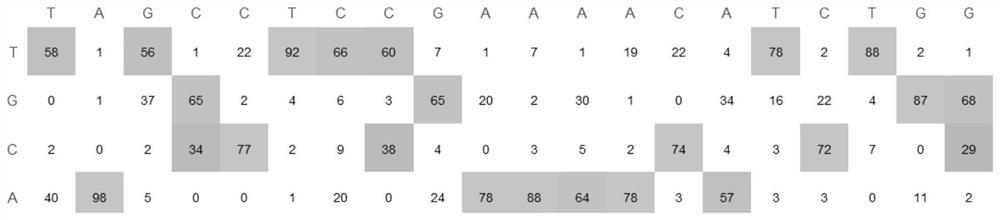
[0090] Example 2, non-integrating lentivirus delivery of ABE to achieve specific site editing
[0091] Using non-integrating lentiviral packaging ABE base editors to harvest virus particles and infect target cells can achieve gene editing at specific sites, which is a relatively safe and efficient method.
[0092] Experimental process: The coding gene of the currently commonly used SpRY ABEmax base editor is constructed into a lentiviral vector. The specific construction method is as follows:
[0093] Step 1: Use the full-length plasmid of the SpRY ABEmax base editor as a template, and use the following primer pair SpRYABEmax-F / SpRY ABEmax-R as amplification primers to perform PCR amplification to obtain the target fragment.
[0094] The specific primer sequences are as follows (lowercase bases are homology arms):
[0095] SpRY ABEmax-F: 5'-aacacaggaccggttctagaATGAAACGGACAGCCGA-3';
[0096] SpRY ABEmax-R: 5'-aagtttgttgcgccggatccGACTTTCCCTCTTCTTCT-3'.
[0097] Step 2: Digest...
Embodiment 3
[0106] Example 3, non-integrated lentiviral delivery PE system realizes editing of specific sites
[0107] It is a relatively safe and efficient method to use non-integrated lentiviral packaging PE system to infect target cells after harvesting virus particles to achieve gene editing at specific sites.
[0108] Experimental process: The coding gene of the currently commonly used PE precision editing system is constructed into a lentiviral vector, and the puromycin gene is expressed in the vector, which can be used for subsequent screening. The specific construction method is as follows:
[0109] Step 1: Use the full-length plasmid of the PE base editor as a template, and use the following primer pairs as amplification primers to perform PCR amplification to obtain the target fragment.
[0110] The specific primer sequences are as follows (lowercase is the homology arm):
[0111] PE-F: 5'-aacacaggaccggttctagaATGAAACGGACAGCCGA-3';
[0112] PE-R: 5'-aagtttgttgcgccggatccGACTTTC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com