Primer pair for identifying eggplant dominant parthenocarpy gene by using InDel molecular marker and application of primer pair
A parthenocarpic and molecular marker technology, applied in the field of molecular biology, can solve the problems of long breeding cycle of parthenocarpic inbred lines and new varieties, great influence of environmental conditions, inaccurate selection, etc. The effect of few limiting factors and high selection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
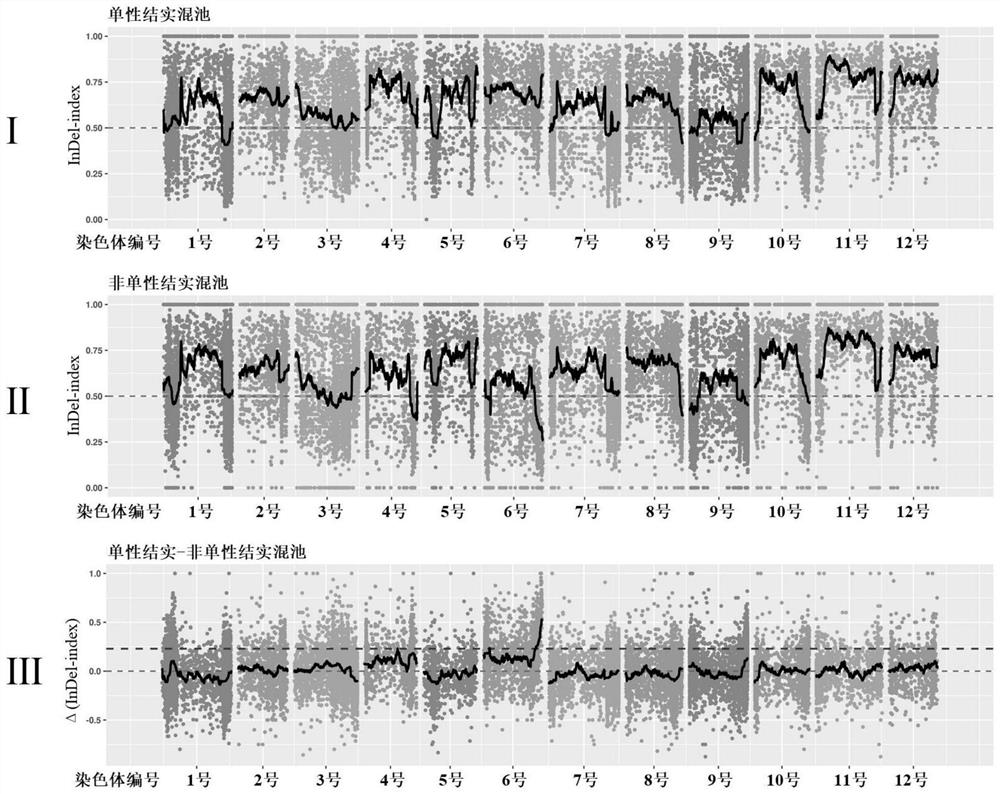
[0062] This embodiment screens the InDel molecular marker linked to the eggplant dominant parthenocarpy gene, and utilizes the InDel molecular marker to identify the eggplant dominant parthenocarpy gene, including the following steps:
[0063] (1) identify parthenocarpy of eggplant plant:
[0064] The test materials are eggplant dominant parthenocarpic inbred line D-10 and non-parthenocarpic inbred line 03-2 independently selected and bred by the eggplant genetics and breeding research group of the Institute of Vegetable and Flower Research, Chinese Academy of Agricultural Sciences, and constructed using them as parents. the F 1 and F 1 F obtained by selfing 2 Segregate groups where 2020 F 2 Population contains 284 individual plants, 2021 F 2 The population contained 392 individual plants. The field trials in 2020 and 2021 were all carried out at the Langfang Experimental Farm of the Vegetable and Flower Research Institute of the Chinese Academy of Agricultural Sciences. ...
Embodiment 2
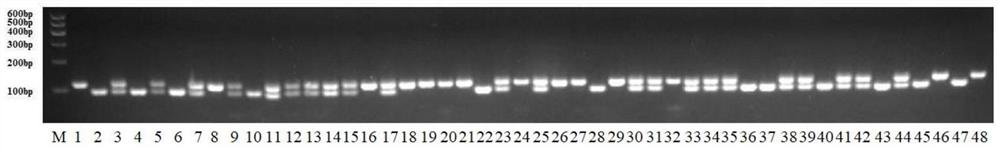
[0093] This example verifies the accuracy of the InDel molecular markers screened in Example 1 in assisted selection of parthenocarpic strains, the steps are as follows:
[0094] The molecular markers for identifying dominant parthenocarpy characteristics obtained in Example 1 were verified by using 92 eggplant materials with different genetic backgrounds. Among the 92 materials, 51 were dominant parthenocarpy materials created by the dominant parthenocarpy line D-10 and different eggplant inbred lines, and 41 were non-parthenocarpic materials. The field phenotype and molecular marker typing results of the test materials were analyzed by PCR amplification. The amplification system and procedure were the same as those in Example 1.
[0095] The results showed that among the 92 materials, the phenotypic data reflected by the molecular markers were inconsistent with the field investigation results in 4 materials, and the accuracy rate was 95.65%.
Embodiment 3
[0097] This embodiment provides a kit for identifying dominant parthenocarpy gene in eggplant, said kit for identifying dominant parthenocarpy gene in eggplant includes a primer pair and a buffer for identifying dominant parthenocarpy gene in eggplant using InDel molecular markers , DNA polymerase, dNTPs, Mg 2+ , Standard Positive Template and Water.
[0098] The nucleotide sequences of the primer pair for identifying eggplant dominant parthenocarpy gene using InDel molecular markers are shown in SEQ ID No.1-2.
[0099] SEQ ID No. 1: GACTACCACTACAATTGTCAT;
[0100] SEQ ID No. 2: TCCAAGCACAACCTCAAGT.
[0101] The buffer, DNA polymerase, dNTPs and Mg 2+ form a mixture;
[0102] The standard positive template is the genomic DNA solution of eggplant dominant parthenocarpic inbred line D-10 with a concentration of 30 ng / μL.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com