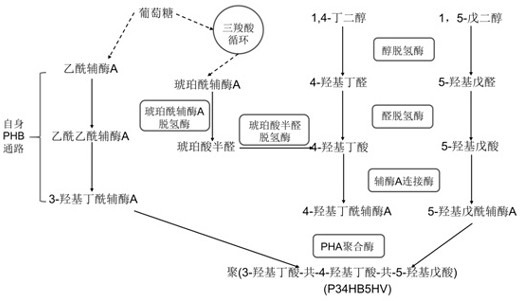
Poly (3-hydroxybutyrate-4-hydroxybutyrate-5-hydroxyvalerate) tripolymer and construction of microbial production strain thereof
A polyhydroxyalkanoate and gene technology, applied in the fields of biotechnology and biomaterials, can solve the problems of insufficient PHA species and limited fields, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0206] transform Halomonas bluephagenesis TD01 CGMCC. No. 4353 for the production of poly(3-hydroxybutyrate-4-hydroxybutyrate-5-hydroxyvaleric acid)
[0207] 1. Plasmid and production strain construction
[0208] 1. Construction of plasmid pY01-pY07
[0209] The backbone of the pY01-pY07 plasmid is pSEVA321 (see Martínez-García E, Aparicio T, Goñi-Moreno A, et al. SEVA 2.0: an update of the Standard European Vector Architecture for de- / re-construction of bacterial functionalities[J] . Nucleicacids research, 2015, 43(D1): D1183-D1189.)
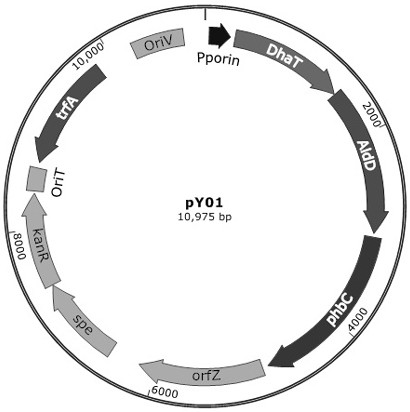
[0210] Preparation of plasmid pY01, the schematic diagram of the structure of plasmid pY01 is shown in Figure 2a As shown, it is a circular plasmid, and its sequence is shown in SEQ ID NO:5.
[0211] Among them, nucleotides 44-261 are porin promoter (GenBank Accession CP007757.1) and its RBS, nucleotides 310-1494 are dhaT gene (GenBank Accession AE015451.2), and nucleotides 1501-3021 The nucleotide is the aldD gene (GenBank Accession AE01...
Embodiment 2
[0287] engineered Halomonas Halomonas campaniensis LS21 CGMCC No.6593 Controlled Production P34HB5HV
[0288] one. Construct production bacteria
[0289] Transform any of the plasmids pY01-pY07 and the pQ08 plasmid (plasmid construction is the same as in Example 1) into Halomonas by conjugative transformation Halomonas campaniensis In LS21 CGMCC No.6593, the obtained Halomonas were named as H. LS21(pY01), H. LS21(pY02), H. LS21(pY03), H. LS21(pY04), H. LS21(pY05), H. LS21(pY06), H. LS21(pY07).
[0290] two. characterize
[0291] 1. Prepare the seed solution.
[0292] 2. Take 2.5mL of the Halomonas seed solution obtained in step 1, inoculate it into a 500mL shake flask containing 47.5mL of shake flask culture medium, and incubate with shaking at 37°C and 200rpm for 48 hours.
[0293] After the shake flask culture was completed, the dry weight of cells, the content of PHA and the content of monomers were measured. Three repeated experiments were carried ou...
Embodiment 3
[0324] After applying the modified Halomonas aydingkolgenesis M1 CGMCC NO.19880 Controlled Production P34HB5HV
[0325] one. Construct production bacteria
[0326] Transform any of the plasmids pY01-pY07 and the pQ08 plasmid (plasmid construction is the same as in Example 1) into Halomonas by conjugative transformation Halomonas aydingkolgenesis In M1 CGMCC NO.19880, the obtained Halomonas were named as H. aydingkolgenesis M1(pY01), H. aydingkolgenesis M1(pY02), H. Aydingkolgenesis M1(pY03), H. aydingkolgenesis M1(pY04), H. aydingkolgenesis M1(pY05), H. aydingkolgenesis M1(pY06), H. aydingkolgenesis M1(pY07).
[0327] two. characterize
[0328] 1. Prepare the seed solution.
[0329] 2. Fermentation
[0330] Take 2.5 mL of the seed liquid obtained in step 1, inoculate it into a 500 mL shake flask containing 47.5 mL of shake flask culture medium, and culture it with shaking at 37°C and 200 rpm for 48 hours.
[0331] After the shake flask culture ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com