Compounds that selectively and effectively inhibit hakai-mediated ubiquitination, as Anti-cancer drugs
A compound, selected technology, applied in the direction of drug combination, active ingredients of heterocyclic compounds, medical preparations containing active ingredients, etc., can solve problems that have not yet been disclosed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0302] Example 1. Synthesis
[0303] Compounds listed throughout this specification can be prepared according to the following general synthetic routes:
[0304]
Embodiment 2
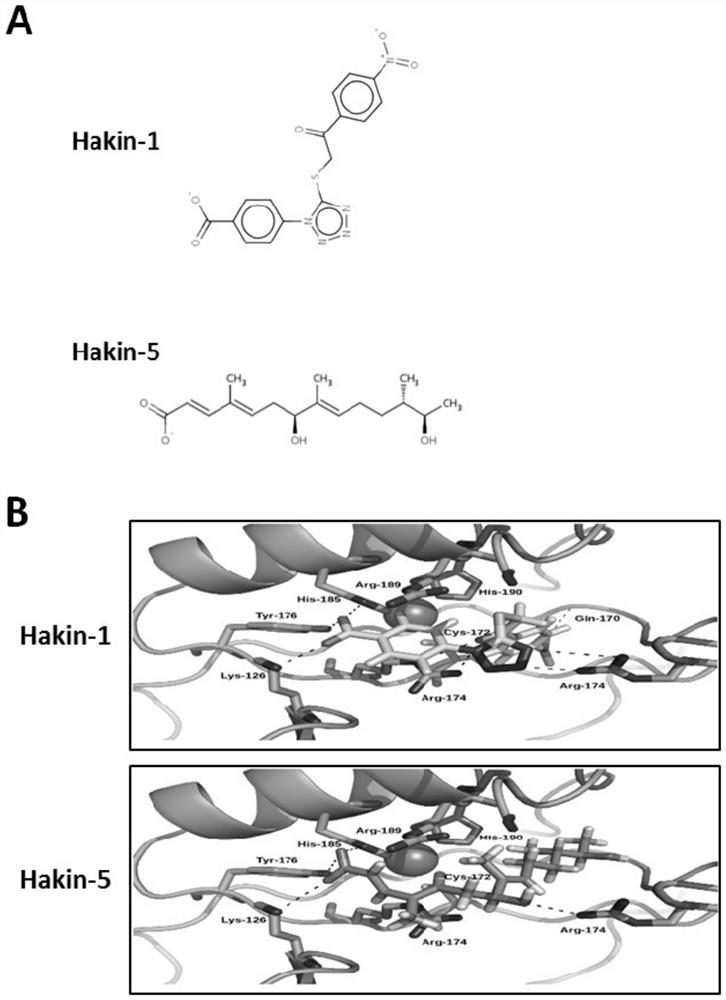
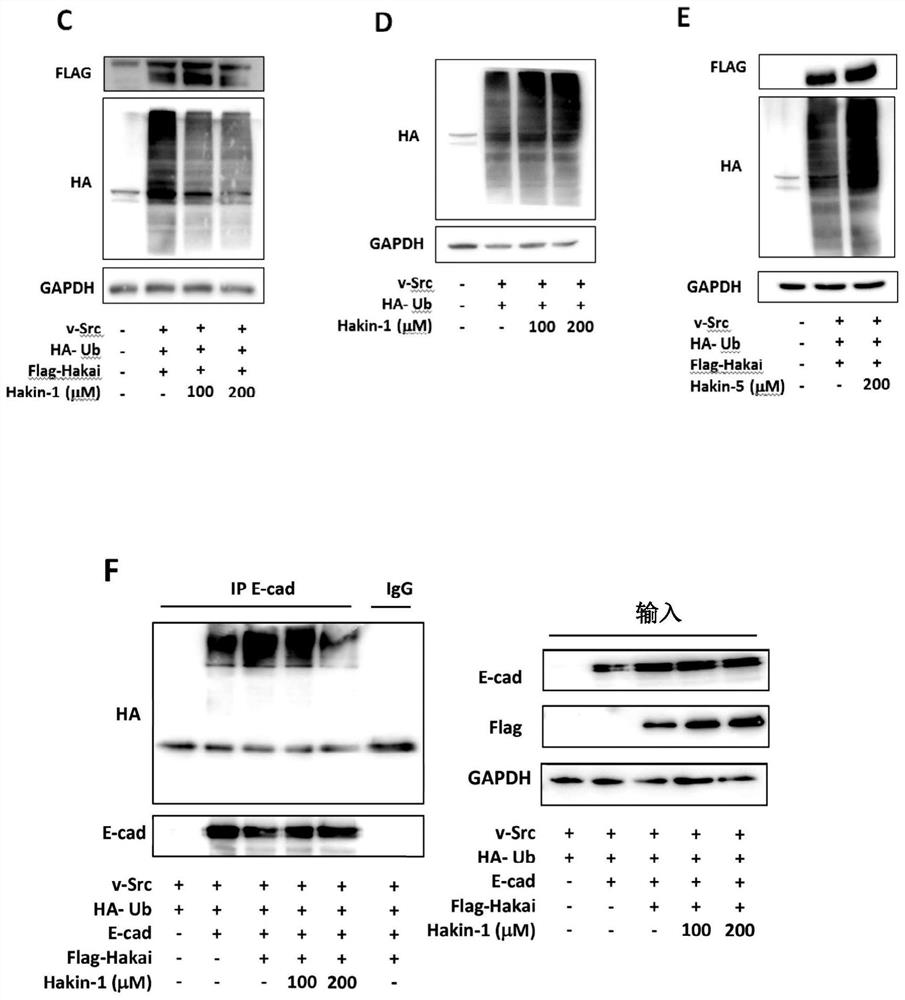
[0306] 2.1. Materials and methods
[0307] Protein and ligand models. The X-ray crystal structure of the phosphotyrosine-binding domain of Hakai (PDB 3VK6) was downloaded from the Protein Data Bank and the dimer was modeled using appropriate symmetry operations. Amino acid protonation was performed at a pH of 7.2 using the pdb2pqr server. 3D models of the ligands were constructed using the Virtual Screening and Data Management Integrated Platform (VSDMIP), as described elsewhere. Briefly, the initial 3D coordinates of each ligand were generated with CORINA [Sadowski, J.; Gasteiger, J.; Klebe, G. Comparison of Automatic Three-Dimensional Model Builders Using 639X-Ray Structures. J. Chem. Inf. Comput. Sci. 1994, 34, 1000-1008 (DOI: 10.1021 / ci00020a039)]. Afterwards, a wide variety of conformers were generated using ALFA [4], and atomic partial charges calculated by MOPAC were assigned to each conformer by employing the AM1 semi-empirical model and the ESP method. All ligand ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com