SNP molecular marker used for identifying Brucella vaccine strain M5, and application of SNP molecular marker
A Brucella and molecular marker technology, applied in the direction of microbial-based methods, recombinant DNA technology, microbial determination/inspection, etc., can solve problems affecting animal brucellosis monitoring and epidemic management, differential diagnosis, human infection, etc. Achieve the effect of simple and efficient identification, rapid detection and typing, and efficient technical support
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Embodiment 1 is used to identify the acquisition of the core SNP molecular marker of Brucella vaccine strain M5
[0059] 1. Systematic analysis of the whole genome information of Brucella using the minimal core component typing method
[0060] (1) Download Brucella melis (136 strains in total) from the GenBank database (http: / / www.ncbi.nlm.nih.gov / genome / ) of the National Center for Biotechnology Information (NCBI) The Complete and Scaffold genomes.
[0061] (2) Based on the Blast method, the gene of M5GCA_000348645.1 was compared with the genomes of 135 strains of Brucella in sequence. If the similarity is greater than 60% and the compared sequence exceeds 70% of the original sequence, the gene is considered to exist in this in the genome. According to this analysis strategy, a total of 1291 Core genes were found in all strains.
[0062] (3) Using the Mummer program, the Core gene sequence was sequentially compared with 134 genomes, the SNP sites were found, and the...
Embodiment 2
[0079] Example 2 Specificity Evaluation of SNP Molecular Marker Detection
[0080]Bartonella henselae, Vibrio cholerae, Francisella tularensis, Escherichia coli O:157, Escherichia coli O:16, Enterocolitica with serological cross-reactivity with Brucella or close relatives to Brucella species Mori bacteria O: 9, Staphylococcus aureus and other kinds of Brucella wild strains, multiple vaccine strains such as S2 (pig species), A19 (cattle species) etc. are used as specific controls as samples to be tested, for example 1 The screened SNP molecular markers and their primer pairs and probe pairs (see Table 1) were evaluated for detection specificity. The specific methods are as follows:
[0081] 1. Genomic DNA extraction
[0082] Bacterial genomic DNA extraction kits from Beijing Tiangen Biochemical Reagent Co., Ltd. were used in strict accordance with the instructions in the kits, including the above-mentioned Bartonella henselae, Vibrio cholerae, Francisella tularensis Escherichi...
Embodiment 3
[0089] The application of embodiment 3 SNP molecular marker in distinguishing Brucella vaccine strain M5 and place Brucella bacterial strain
[0090] Utilize the SNP molecular marker that screening obtains in embodiment 1 and its primer (SEQ ID NO.2-3) and probe (SEQ ID NO.4-5) to Brucella vaccine strain M5, Brucella standard strain, 120 Brucella wild strain isolated from local strains and Brucella vaccine strain S2 (pig species), A19 (cattle species) are differentiated and identified, and the extraction of genomic DNA and the RT-PCR method are the same as in Example 2.
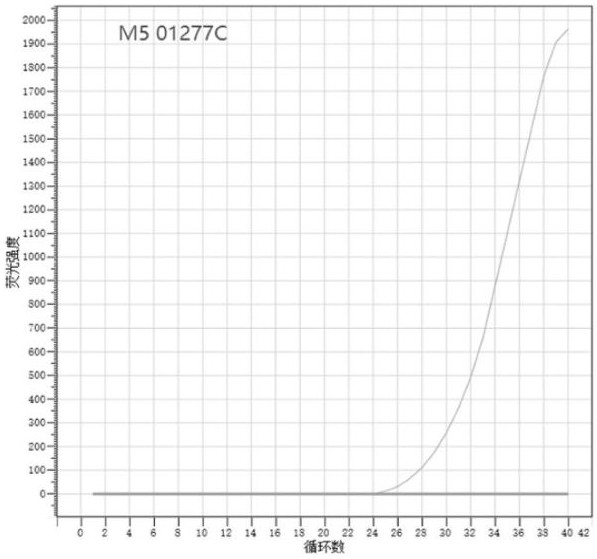
[0091] The identification results are shown in Table 2, Table 3 and Table 4. The fluorescence signal of the amplification curve of Brucella vaccine strain M5 is the fluorescence (FAM) labeled with the probe shown in SEQ ID NO.4, and the corresponding SNP site is A, The fluorescence signal of the amplification curve of each standard strain (see Table 4 for the name of the standard strain) is the fluorescence (...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com