Molecular marker co-separated from watermelon peel covering line gene ClGS and application of molecular marker
A molecular labeling and co-separation technology, which is used in the determination/inspection of microorganisms, biochemical equipment and methods, food processing, etc. handy effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
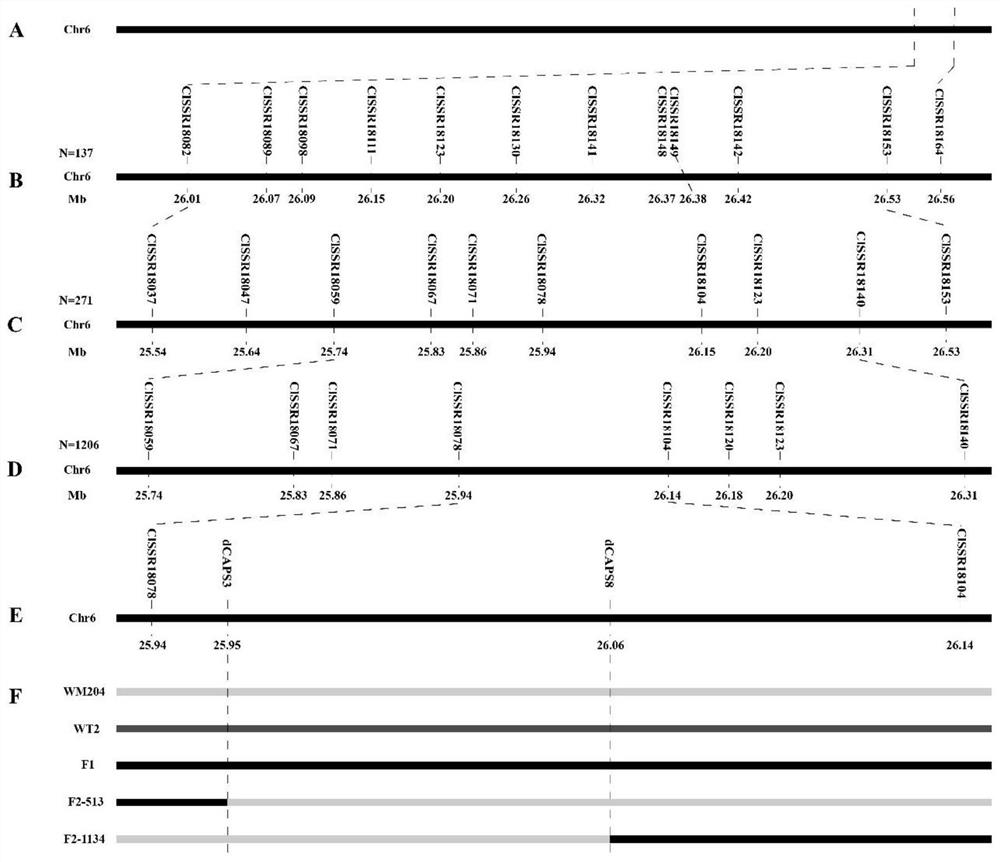
[0030] Example 1 Acquisition of Molecular Markers Closely Linked to Watermelon Plant Dark Green Ribbon Gene
[0031] biomaterials:
[0032] Watermelon material WM204, its peel is light green, covered with net pattern;
[0033] The watermelon material WT2 is a high-generation inbred line selected by the inventor. The fruit is light green peel and covered with dark green band-like patterns on the fruit surface. This material can be obtained through commercial channels or obtained from Henan Agricultural University. Provided by the Genetics and Breeding Research Group (it needs to be explained that the use of this material as a research basis is only for the convenience of obtaining experimental materials, and it should not be understood that the realization of the relevant technical solutions of this application must rely on this experimental material);
[0034]The above-mentioned watermelon material WT2 is consistent with the normal watermelon material WT2 with tendrils used i...
Embodiment 2
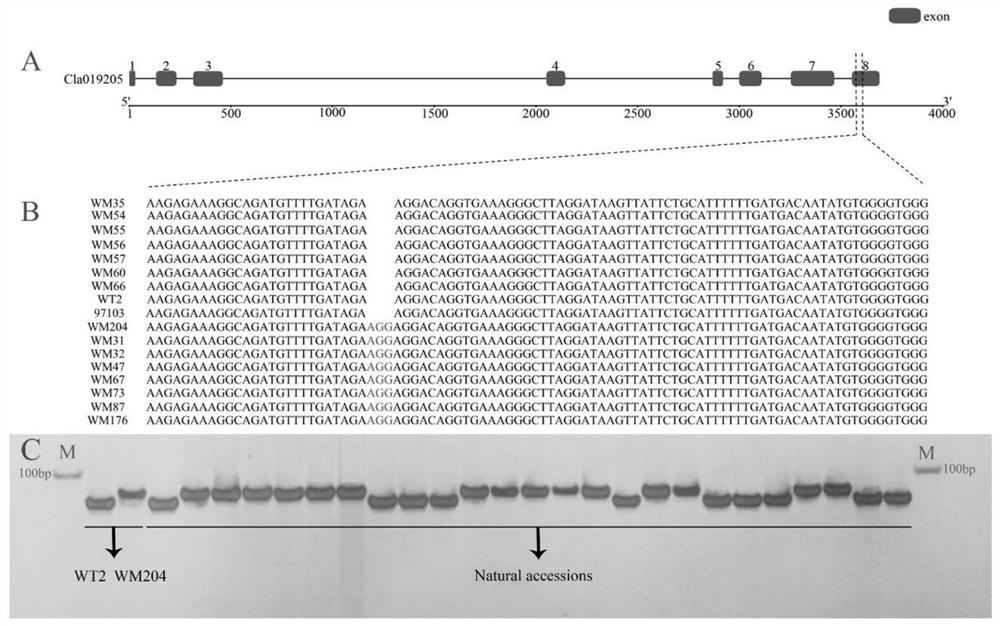
[0071] Example 2 Development of Indel1 marker
[0072] On the basis of Example 1, the inventors further performed a bioinformatics analysis on the candidate segment of the watermelon dark green band gene ClGS, and developed an Indel1 marker co-segregated with it. The specific process is as follows.
[0073] (1) Parental resequencing sequence analysis
[0074] On the basis of the fine mapping in Example 1, the inventors further conducted a bioinformatics analysis on the candidate segment of the watermelon dark green banded pattern gene ClGS. We took the genome sequence between dCAPS3 and dCAPS8 as the reference sequence, and according to the annotation of the reference genome, there were 11 genes in the candidate interval. Then, we mapped the original read length on the reference sequence, and analyzed it through software such as hisat2 and samtools. We found a total of 64 SNPs and 3 Indel sites in the CDS.
[0075] (2) Electronic BSA
[0076] On the basis of step (1), in o...
Embodiment 3
[0088] Example 3 Indel1 molecular marker function verification
[0089] On the basis of the above-mentioned embodiments, based on the collected watermelon germplasm materials from all over the world, the inventor randomly selects 25 parts of materials as an example (and the watermelon germplasm materials are all collected during the inventor's work, relevant Germplasm materials are also preserved in some domestic and foreign professional germplasm banks, and the following work is only for experimental verification, and the relevant germplasm materials are not directly related to the protection subject of this application, so these germplasm materials are no longer provided information), the Indel1 molecular marker function was further verified, and the specific process is briefly introduced as follows.
[0090] (1) Extract watermelon genomic DNA
[0091] Genomic DNA was extracted from unexpanded young leaves after seedlings (collected from the parental dark green banded-patte...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com