Molecular marker AhyRscc closely linked with deep red seed coats of peanuts and application of molecular marker AhyRscc
A molecular marker, deep red technology, applied in the field of agricultural biology, can solve the problems of peanut variety restrictions, achieve reliable results, improve breeding efficiency, and simple technical requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
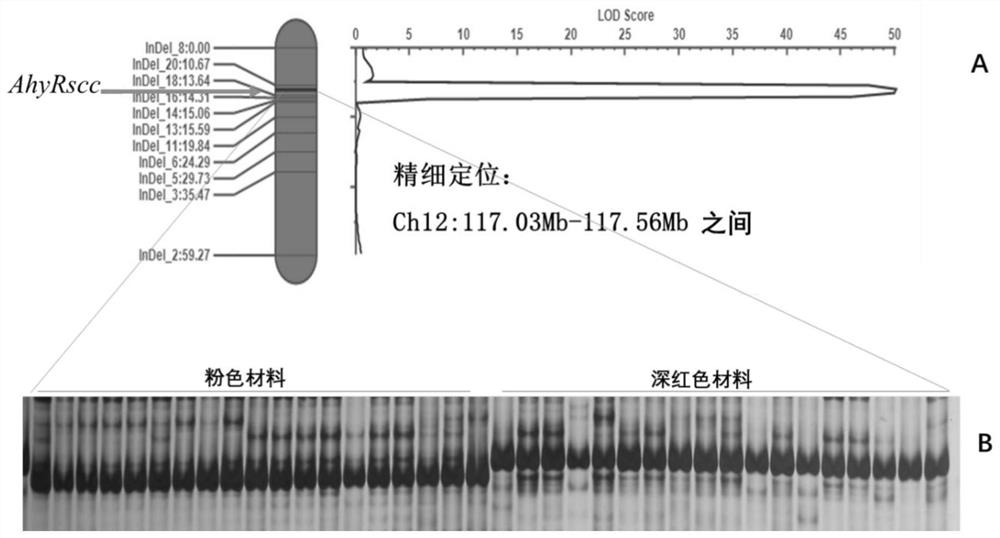
[0035] Example 1 Controlling the positioning of the peanut crimson seed coat color gene and the design of the molecular marker AhyRscc closely linked with the peanut crimson seed coat
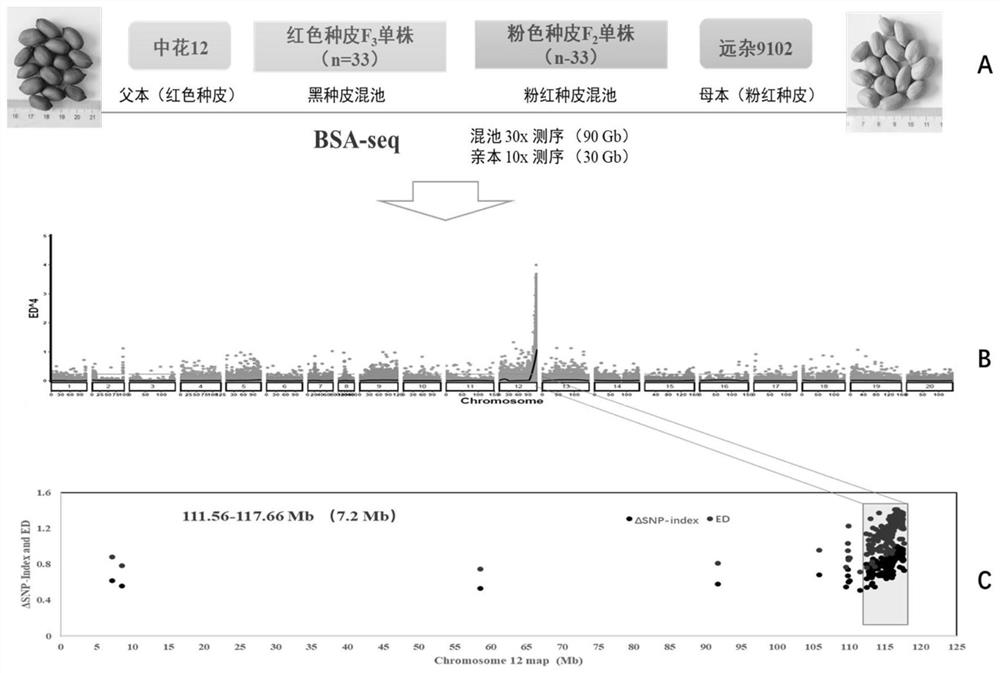
[0036] In order to locate and control the color gene of peanut crimson seed coat, the inventors used the pink seed coat peanut variety (female parent) Yuanza 9102 to cross with the deep red seed coat peanut variety (male parent) Zhonghua 12 to construct a segregation population, genetic Analysis reveals that seed coat color in peanuts is controlled by a single gene.
[0037] by F 2 and F 3 For the phenotype analysis of the generation, 33 extreme red and 33 extreme pink materials were selected to construct the deep red and pink extreme pools respectively, and the whole genome resequencing was carried out together with the parents Zhonghua 12 and Yuanza 9102, and the parents and extreme The sequencing volumes of the pools are 30Gb and 90Gb respectively ( figure 1A). The resequencing data were...
Embodiment 2
[0039] Example 2 Using the molecular marker AhyRscc to rapidly breed a new high-yield crimson peanut variety
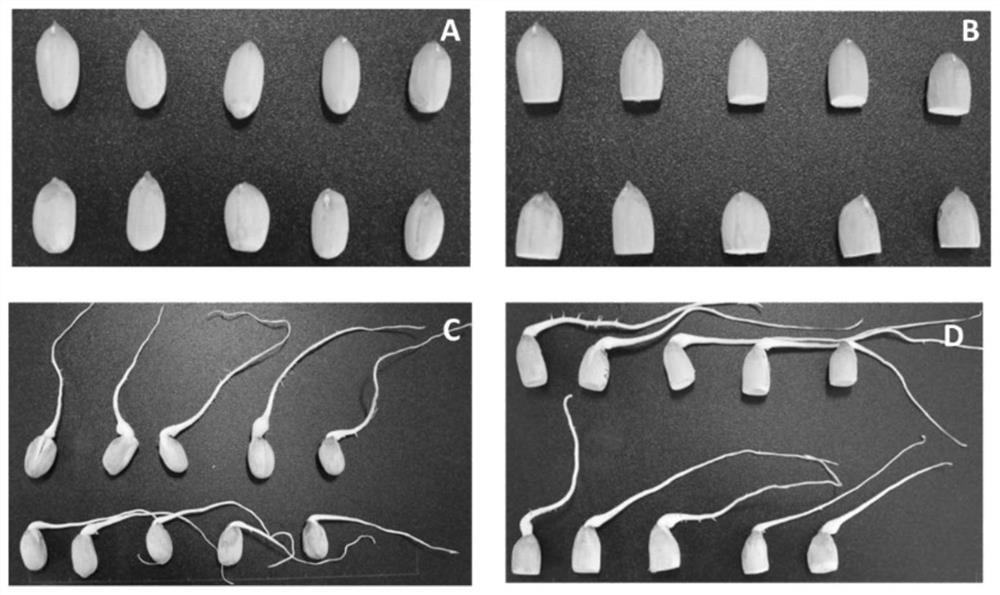
[0040] By using the molecular marker AhyRscc for selection combined with backcross breeding, common peanuts (recurrent parents) can be improved into new varieties of peanuts with dark red seed coats in about 3 years, and more than 97% of the genetic background of the recurrent parents can be retained. On the premise of retaining most of the excellent traits of the original peanut varieties, the directional improvement of the color of the seed coat from pink to deep red is realized.
[0041] The common peanut variety Fenghua No. 1 was used as the recurrent parent, and the deep red peanut variety Zhonghua No. 12 was used as the donor male parent. The specific steps for improving the color of the peanut dark red seed coat include:
[0042] (1) hybridization
[0043] HF001 was used as the female parent (reincarnation parent) and the dark red peanut variety Zhonghua No. 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com