Porcine fibroblast line with double sgRNA, gene knockout vector and gene knockout STING gene and construction method of porcine fibroblast line
A gene knockout and carrier technology, applied in the field of porcine fibroblast cell lines and its construction, can solve problems such as the inability to guarantee the automatic repair of cells, the inability of cell lines to be cloned, and the risk of uncertainty in cloning operations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Construction of CRISPR / Cas9 targeting vector targeting STING gene
[0058] 1. Use the NCBI database to search the gene sequence of porcine STING (Gene ID: 100217389), and locate the first exon (fourth exon) and the last exon ( Exon 8) segment for target design.
[0059] The sgRNA was designed using the website http: / / crispor.tefor.net / . The sgRNA fragments targeting the fourth exon were named: STING-sgRNA1, STING-sgRNA2, STING-sgRNA3; the sgRNA fragments targeting the eighth exon were named: STING-sgRNA4, STING-sgRNA5. Add cohesive ends to both ends of the sgRNA according to the BbsI restriction endonuclease. The method is: add the base CACCG at the 5' end of the sgRNA sequence to obtain a forward fragment; if the first base at the 5' end of the sgRNA sequence is not G, then Add a CAAA base sequence to the 3' end of the sgRNA reverse complementary strand and add a C base to the 5' end as a reverse fragment; if the first base at the 5' end of the sgRNA sequence is G, t...
Embodiment 2
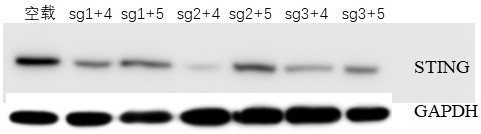
[0101] Construction of porcine fibroblast cell line with gene knockout of STING gene
[0102] (1) The above plasmids with high cutting efficiency (the expression vector group screened in Example 1, PX459-STING-sgRNA2+PX459-STING-sgRNA4 group). The cells were re-transfected by liposome transfection, and 24 h after transfection, the transfected cells were treated with 1.5 μg / mL puromycin for 3 days. After two rounds of selection, all the cells in the control group died. After the cells in the experimental group were digested, monoclonal cells were picked by the limiting dilution method and expanded for culture.
[0103] (2) Identify all the mononuclear clonal cells picked, and extract the genomic DNA of the cells for PCR identification. The primers used for PCR identification were STING-F1: 5'-TGGATTTCTTGGTTCCTACAG-3' (nucleotide sequence No. 11) and STING-R1: 5'-CGAGTCCTCACCCTGGTAGA-3' (nucleotide sequence No. 12).
[0104] The amplification conditions were 95°C, 5min; 95°C, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com