Method for knocking out saccharomyces cerevisiae chromosome
A technology for Saccharomyces cerevisiae and Saccharomyces cerevisiae strains, applied in other methods of inserting foreign genetic materials, stably introducing foreign DNA into chromosomes, biochemical equipment and methods, etc. Complete chromosomes have a large workload and other problems, to avoid crossovers, complete knockouts, and high success rates
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
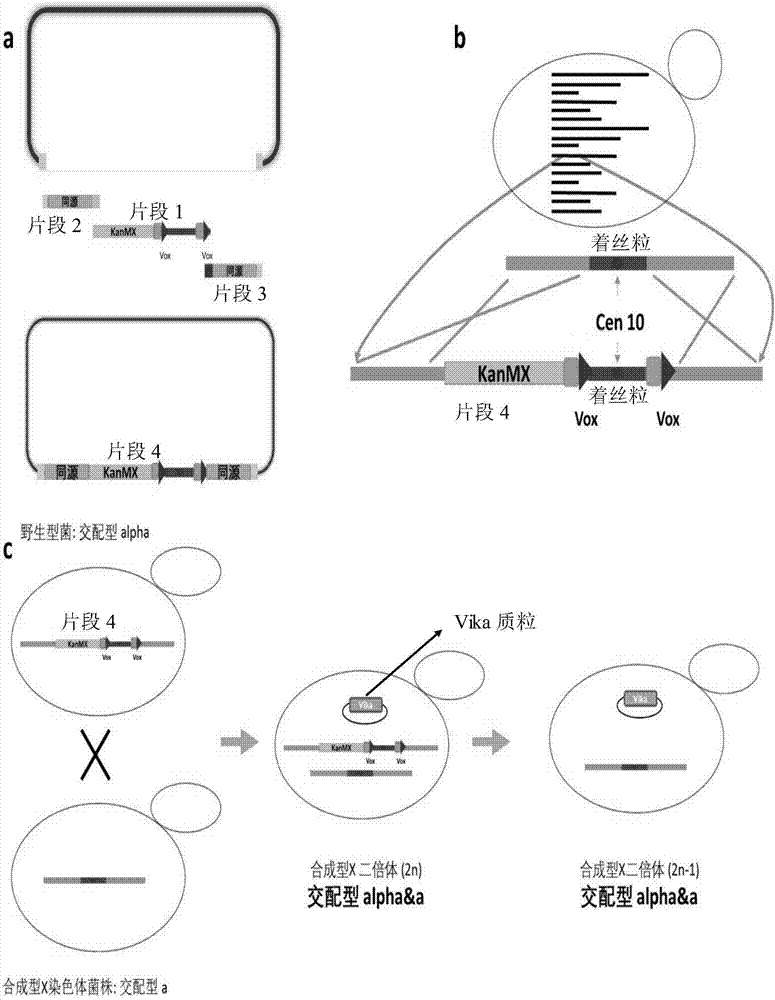
[0032] Example 1: Knockout of chromosome X to be knocked out in commercial BY4742 Saccharomyces cerevisiae
[0033] 1. Fragment construction
[0034] Design upstream and downstream primers to PCR amplify and synthesize the KanMX resistance gene, and the gene between the two vox sites (20bp homologous KanMX resistance gene + vox + chromosome X chromosome sequence + vox), the left end of the gene between the two vox sites (upstream of vox) has 20bp homology with KanMX resistance gene;
[0035] Extract the genome of BY4742 strain, design the upstream primer with 20bp pUC19 plasmid EcoRI restriction site upstream homology and the right end with 20bp KanMX resistance gene homology sequence on the left end, and the downstream primer right end with 20bp pUC19 plasmid HindIII restriction site downstream homology and left end band 20bp vox homologous sequence, PCR amplified 1000bp homologous sequence at both ends of chromosome X chromosome of wild type BY4742 Saccharomyces cerevisiae;...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com