crRNA combination for multiple detection of hereditary hearing loss, kit and method theref
A genetic deafness, multiple detection technology, applied in the direction of recombinant DNA technology, DNA / RNA fragments, biochemical equipment and methods, etc., can solve the problems of unfavorable large-scale genetic screening, limitation, low throughput, etc., to avoid Effects of aerosol pollution, cost reduction, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
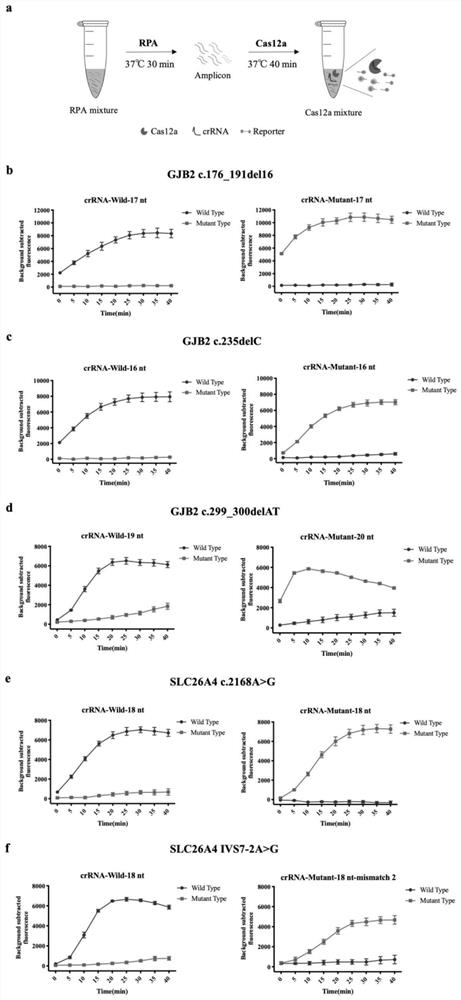
[0064] Example 1: Screening the most suitable crRNA based on Cas12a two-step method
[0065]According to literature reports, the crRNA of CRISPR / Cas12a is composed of Direct repeat and Spacer. Direct repeat mainly plays the role of anchoring Cas protein, and Spacer mainly plays the role of recognizing template DNA, and Spacer of different lengths can recognize and distinguish mutation sites for Cas12a. The efficiency of the point has a great influence (Zetsche B, et al., 2015, Cell, 163(3): 759–771; Dong D, et al., 2016, Nature, 532(7600): 522–526; Li SY, et al., 2018, Cell Discov, 4:20). Therefore, we first designed crRNAs of different lengths according to the GJB2 gene mutation sites c.176-191del16, c.235delC, c.299_300delAT and SLC26A4 gene mutation sites c.2168A>G, IVS7-2A>G, where the Direct repeat sequence change the length of the Spacer sequence (15-24nt); at the same time, for the crRNA with a high signal-to-noise ratio, it is further improved by introducing a mutat...
Embodiment 2
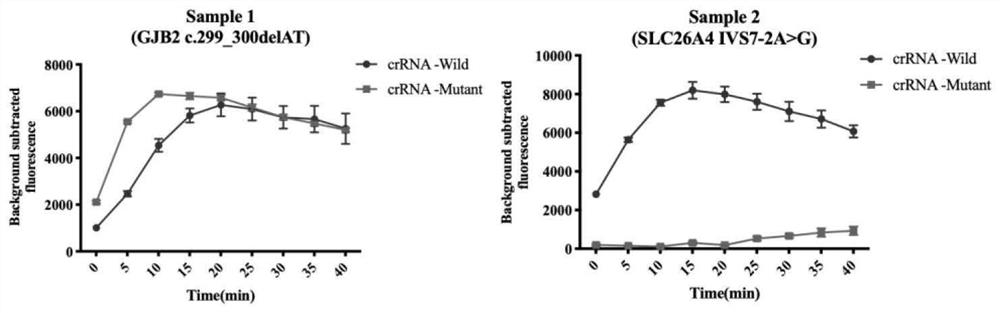
[0077] Example 2: Detection and typing of deafness genome samples
[0078] The most suitable crRNA screened in Example 1 was used for the detection of deafness genome samples. The detection method was the same as in Example 1. Each system was added with 1 μL of the genome sample to be tested, and each reaction was set for 3 repetitions. The detection results were as follows: figure 2 shown. Depend on figure 2 The results show that when sample 1 is detected with crRNA-Wild-19nt and crRNA-Mutant-20nt at the c.299_300delAT site, the fluorescence curve increases significantly, indicating that the crRNA can complete complementary pairing with genomic DNA, thereby activating Cas12a to cut ssDNA The activity of the reporter proves that the sample contains the GJB2 (c.299_300delAT) mutation site, and it is a heterozygous mutant type; and when the crRNA-Wild-18nt and crRNA-Muatnt-18nt-mis of the IVS7-2A>G site are used 2 When testing sample 2, the fluorescence curve correspondin...
Embodiment 3
[0079] Embodiment 3: Based on Cas12a one-step method test the effectiveness of multiple detection
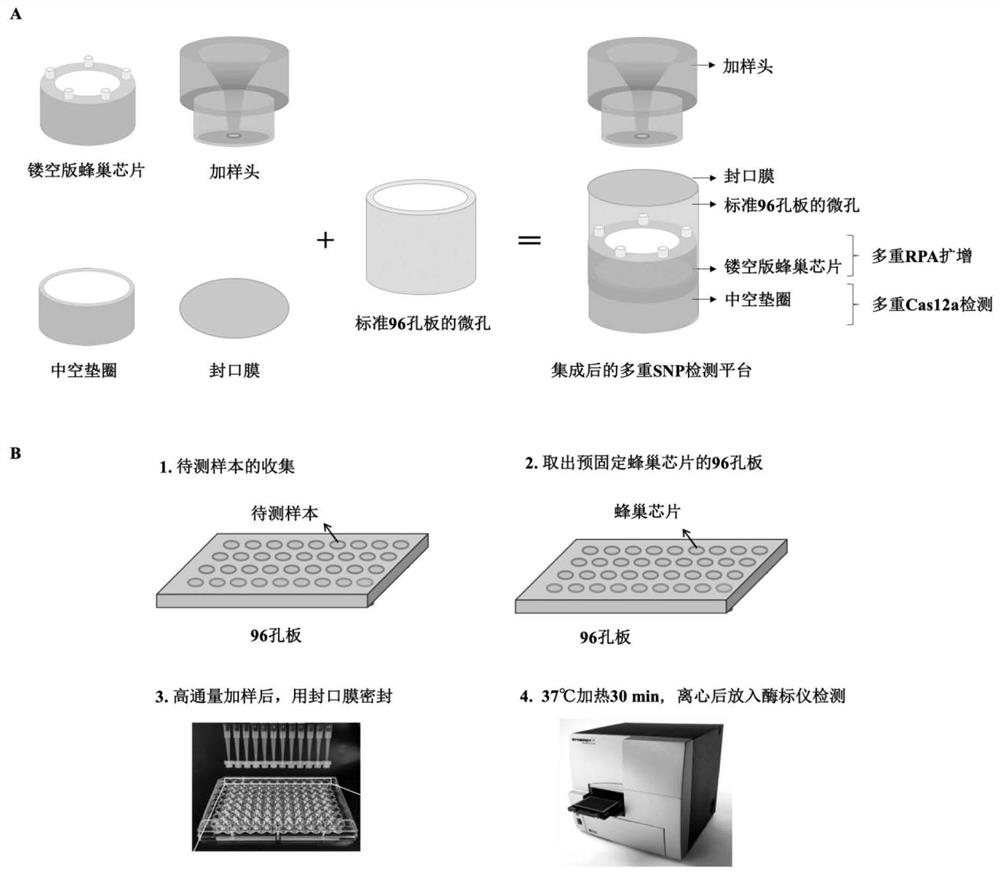
[0080] Since embodiments 1 and 2 have demonstrated the effectiveness of crRNA single detection, this embodiment will be based on the hollow honeycomb chip platform (such as image 3 shown) to test the effectiveness of the Cas12a one-step method for multiplexing.
[0081] The kit structure used is as follows Figure 4-Figure 7 As shown, including hollow honeycomb chip, hollow gasket 3, orifice plate 8 and sampling head 5;
[0082] The hollow gasket 3 is arranged under the hollow honeycomb chip, and the hollow gasket 3 and the hollow honeycomb chip are placed in the microhole 4 of the orifice plate 8;
[0083] The hollow honeycomb chip includes a chip base 2 and a plurality of capillaries 1, the chip base 2 is a hollow ring structure, and the plurality of capillaries 1 are evenly arranged along the circumferential direction of the chip base 2; the upper ends of each capillary ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com