Chili capsaicin content close linkage SNP site, CAPS molecular marker and application thereof
A technology of molecular markers and capsaicin, which is applied in the field of pepper genetics and molecular biology, can solve problems such as genetic complexity, and achieve the effect of improving identification efficiency, identification efficiency and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
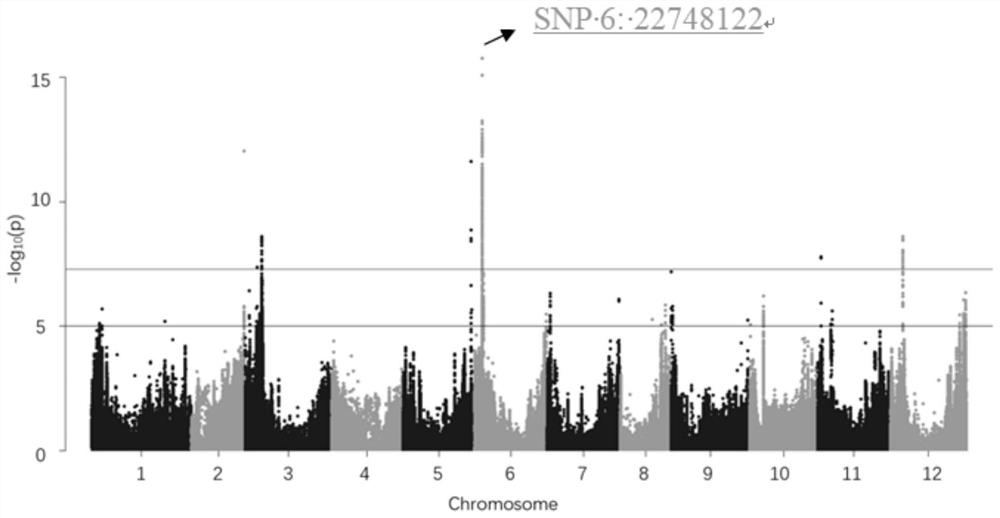
[0036] Example 1 Exploitation and confirmation of closely linked SNP sites for capsaicin content in capsicum
[0037] 1. Acquisition of closely linked SNP sites for capsaicin content in capsicum
[0038] 311 annual capsicum (Capsicum annuum L.) were resequenced. The sequencing platform was IlluminaSolexa. Trimmomatic v0.33 was used to process the raw data to remove adapters, poly-N and low-quality fragments to obtain clean data. Clean data was obtained using BWA0.75 The data was compared to the Zunla-1 genome data (Qin et al., 2014), and the setting parameters were aln-o 1-e 10-t 4-l 32-i 15-q 10; using Samtools 0.1.19 (Li et al. , 2009), using the Bayesian algorithm, setting the parameter -q 1 -C50-S-D-m 2-F 0.002-u, running the command mpileup, searching for SNPs, filtering SNPs, setting the minimum gene allele frequency to be greater than 0.01, and the deletion rate to be less than 0.1.
[0039] 311 annual capsicum (Capsicum annuum L.) fruits were removed from the seeds an...
Embodiment 2
[0047] Example 2 Development of CAPS molecular markers closely linked to capsaicin content
[0048] 1. Design of CAPS primers
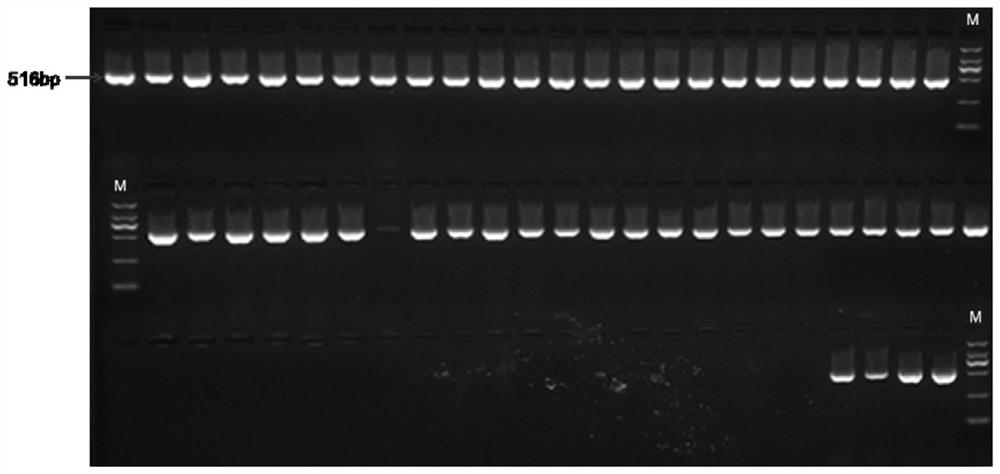
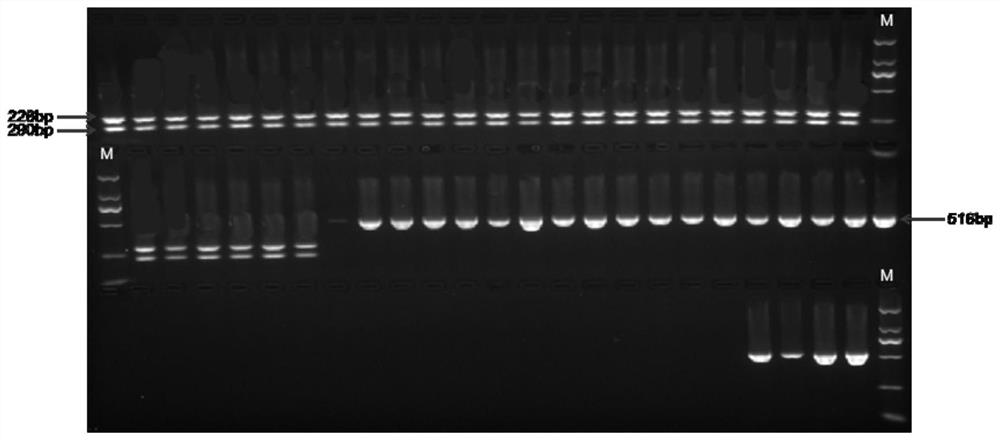
[0049] Design CAPS primers according to the SNP site provided in Example 1, the front primer is CA-capsF: AAAAGGGTTCTAATTCGCGGC (see SEQ ID NO: 2), the back primer is CA-capsR: GGAACTCTGGCTTGCAGGAT (see SEQ ID NO: 3), PCR amplification The product is 516bp, and its sequence is shown in SEQ ID NO: 1 in the sequence listing, and the S at the 227th position of the sequence is G / C, which is a polymorphic site.
[0050] 2. Extraction of sample genomic DNA and PCR amplification
[0051] The young leaves of 26 capsicum materials were selected for genomic DNA extraction for follow-up experiments, including 15 low-capsaicin materials (capsaicin content is less than 1200mg / kg, genotype CC), 11 high-capsaicin materials (capsaicin content The element content is higher than 1200mg / kg, and the genotype is GG). Two plants were taken from each material for paralle...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com