Tandem mass spectrometric identification method for protein based on matching between characteristic information of target database and decoy database
A feature information, two-stage mass spectrometry technology, applied in the field of protein two-stage mass spectrometry identification, can solve the problems of neglecting matching characteristics, affecting the efficiency and accuracy of database search, and errors of biological mass spectrometry instruments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0101] The present invention will be described in further detail below in conjunction with the embodiments and accompanying drawings.
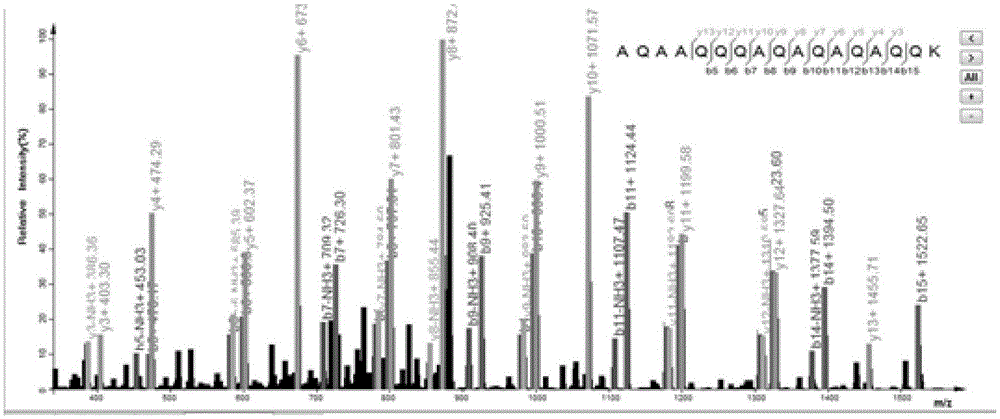
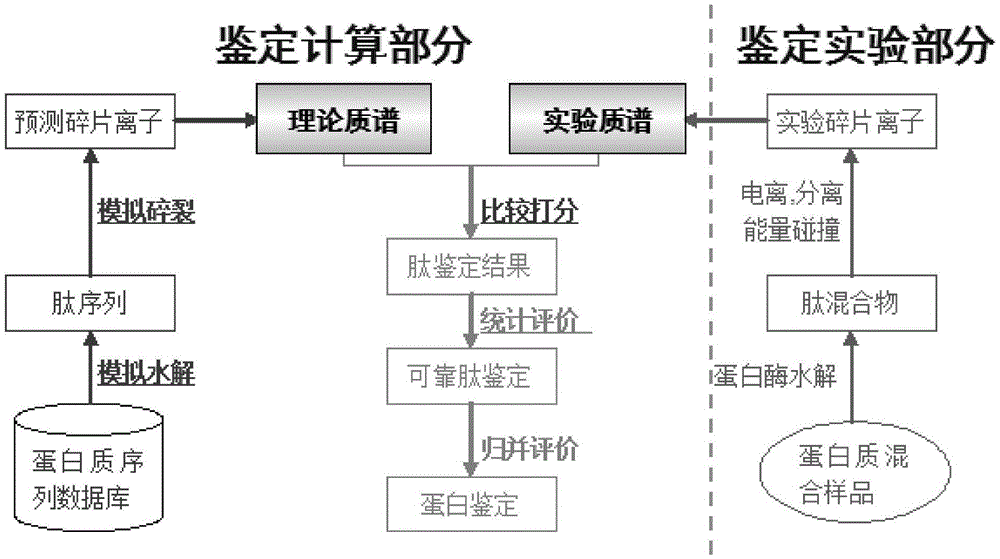
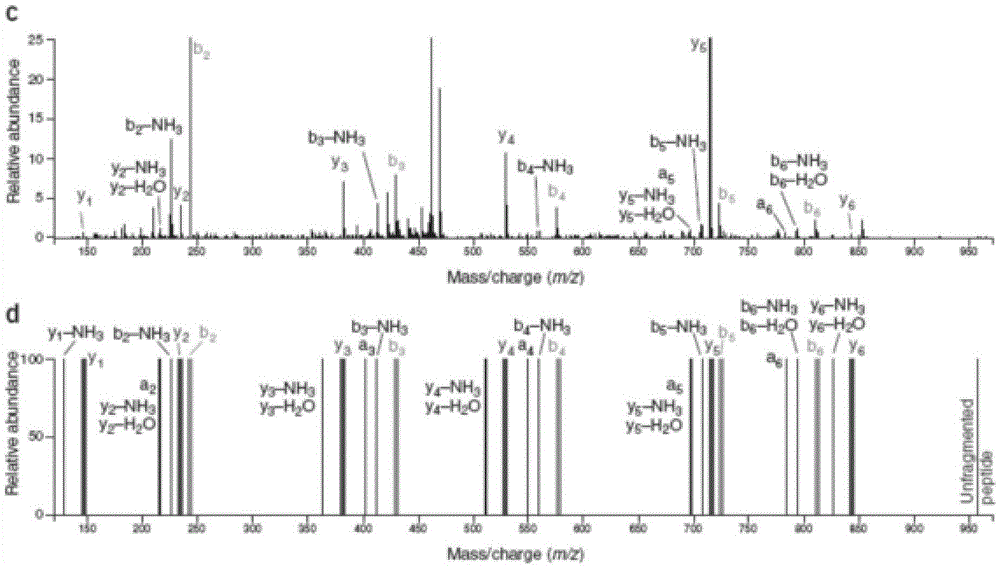
[0102] see Figure 4 As shown, in this embodiment, a protein secondary mass spectrometry identification method based on the matching of positive and negative library feature information includes the following steps:
[0103] (1) Download the protein reference sequence database, and reverse the protein reference sequence to obtain the protein sequence database including the positive library and the reverse library;
[0104] (2) virtual enzymolysis of the above-mentioned protein database sequence, and establish a peptide quality database and a peptide quality database index according to the mass number of the peptide after enzymolysis;
[0105] (3) Remove isotope peaks from the experimental spectrum to be analyzed, and select effective peaks reasonably to improve the signal-to-noise ratio of the experimental spectrum itself;
[0106] (4) Accor...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com