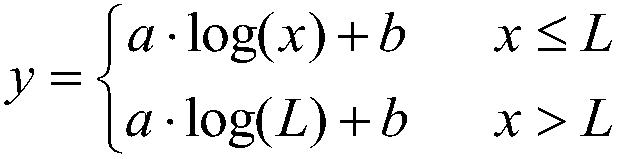Identification method of protein by MS/MS based on multi-omics abundance information
An identification method and secondary mass spectrometry technology, applied in scientific instruments, instruments, measuring devices, etc., can solve the problem of low spectrum ratio, and achieve the effect of saving search space.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
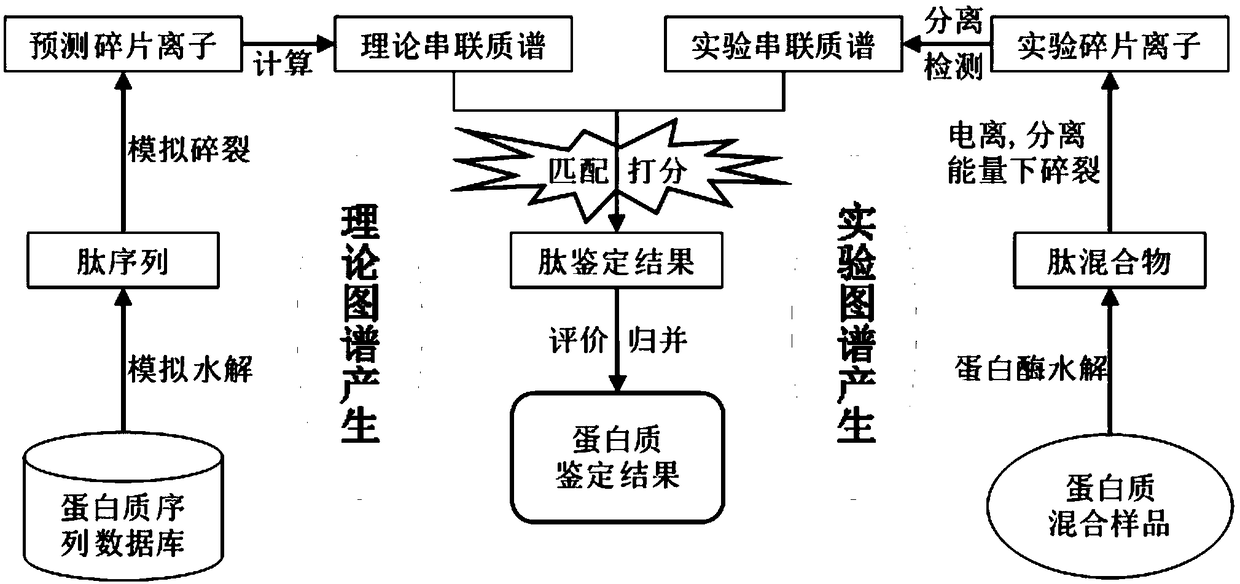
[0052] In order to facilitate the understanding of the present invention, the present invention will be described more fully below with reference to the associated drawings. Preferred embodiments of the invention are shown in the accompanying drawings. However, the present invention can be embodied in many different forms and is not limited to the embodiments described herein. On the contrary, these embodiments are provided to make the understanding of the disclosure of the present invention more thorough and comprehensive.
[0053] This embodiment relates to a method for identifying proteins by MS / MS based on multi-omics abundance information.
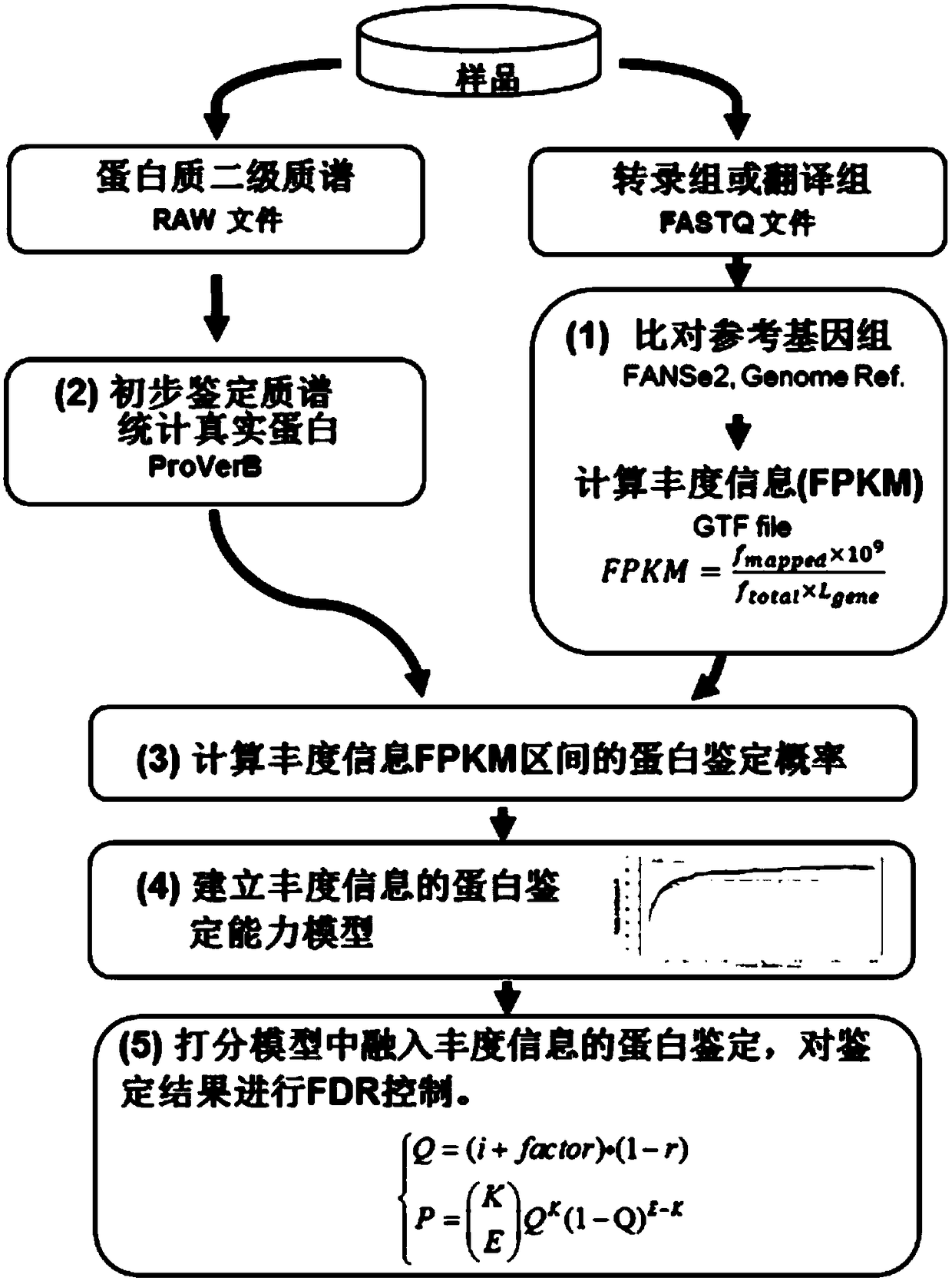
[0054] see figure 2 As shown, the protein secondary mass spectrometry identification method based on multi-omics abundance information includes the following steps:
[0055] (1) The next-generation sequencing base sequence of the transcriptome or translation genome is mapped to the reference genome, and the abundance information o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com