Primer probe combination and kit for detecting pathogen nucleic acids of human herpes virus and application of primer probe combination and kit
A human herpes virus and detection kit technology, applied in the field of microorganism and nucleic acid genome detection, can solve the problems of poor DNA polymerase activity, improper temperature, and invalid indication results, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] The construction of embodiment 1 recombinant plasmid and internal reference plasmid
[0047] The genome sequences of CMV and Human B-globin (HHB) were downloaded from the NCBI website, and the highly conserved sequences were screened and compared with Vector11.1 software.
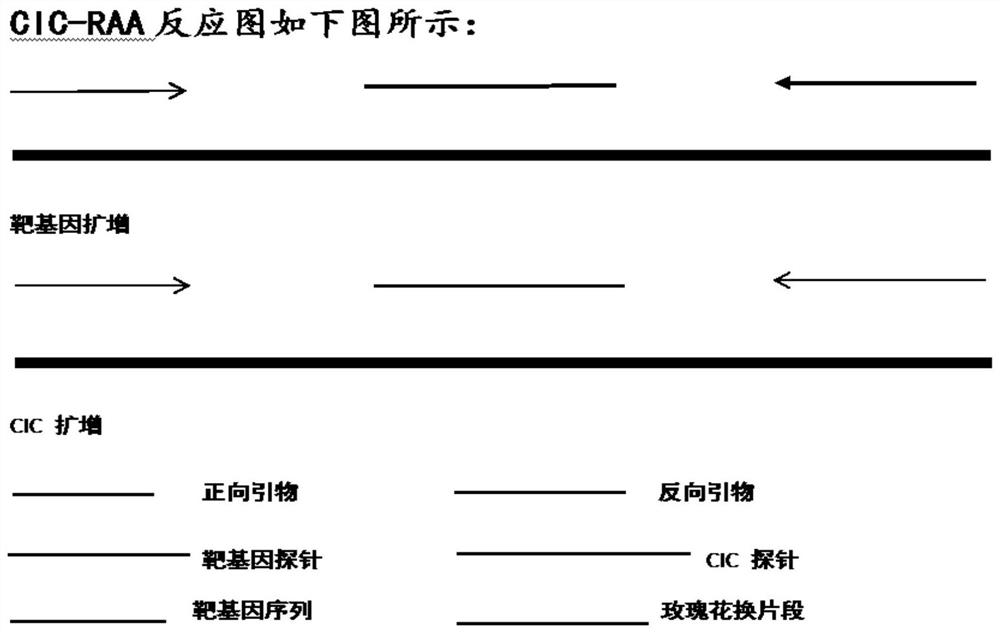
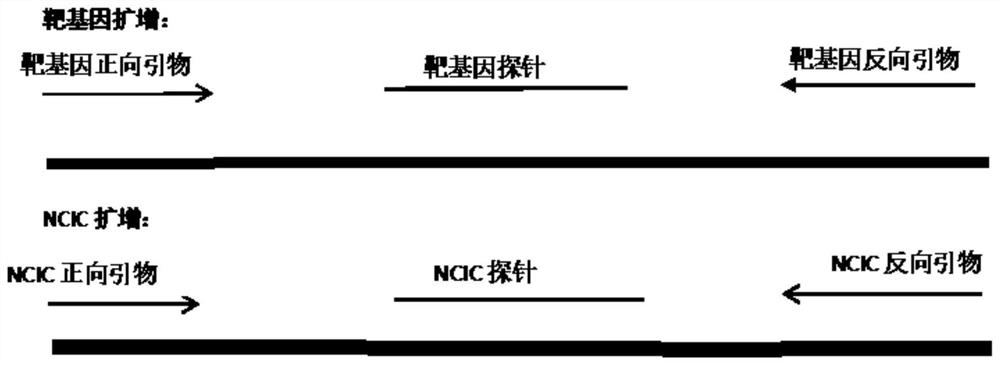
[0048] The CIC sequence replaces the position of the target probe with the rosette sequence in the selected CMV genome sequence, and the CIC probe sequence is a complementary rosette sequence. The position of the CIC probe sequence change and the CIC-RAA reaction diagram of the whole real-time internal control control are as follows figure 1 As shown, the NCIC-RAA response graph is shown as figure 2 shown.
[0049] Entrust a professional company to synthesize and return the target fragment.
Embodiment 2
[0050] Design and synthesis of embodiment 2 primers and probes
[0051] According to the principles of RAA primer and probe design, primers and probes were manually designed for CMV UL123 gene and HHB gene. The lengths of primers and probes ranged from 30-35bp and 46-52bp, respectively. The probes consist of upstream oligonucleotides carrying 5'-FAM (target gene probe) and 5'-HEX (CIC probe), respectively connected to adjacent downstream oligonucleotides via THF spacers. The 3' end of the downstream oligonucleotide has a C3' spacer (polymerase extension blocking group), and the specificity of the primers and probes is analyzed and evaluated with Oligo7.0 software and Primer BLAST software of NCBI. The sequences of the probes are shown in Table 1.
[0052] Table 1
[0053]
[0054]
[0055] All primers and probes were synthesized by a professional company, the primers were purified by PAGE, and the probes were purified by HPLC. The three probes use double-labeled prob...
Embodiment 3
[0056] Example 3 Optimization of RAA
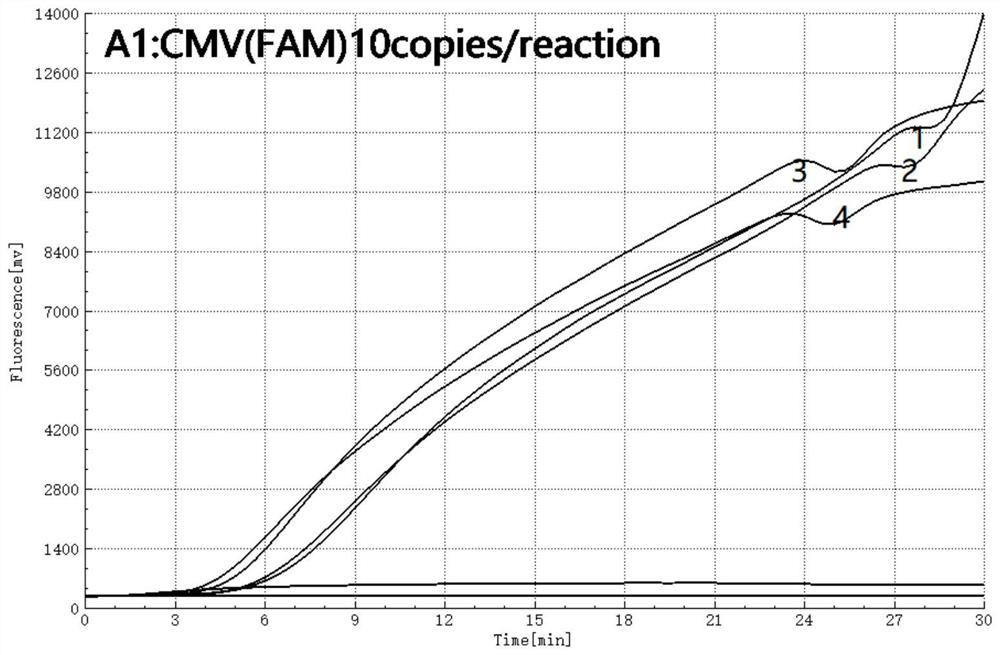
[0057] 1. Optimization of CIC-RAA: In order to determine the optimal concentration and template concentration of CIC, and ensure that the normal amplification of CIC does not affect the target gene amplification curve, different CIC template concentrations (500, 200, 100 and 50 copies / μL ) in the presence of , we optimized the concentration of the CIC template by detecting standard plasmids 1, 5, and 10 copies.
[0058] 1) The reaction system includes: 25 μL buffer, 2.1 μL forward primer (as shown in SEQ ID No.4, 10 μmol / L), 2.1 μL reverse primer (as shown in SEQ ID No.5, 10 μmol / L), 0.6 μL target gene probe (as shown in SEQ ID No.6, 10 μmol / L), 0.6 μL CIC probe (as shown in SEQ ID No.7, 10 μmol / L), 1 μL CIC plasmid (respectively 500, 200 , 100, 50 copies / μL). Adjustable volumes of nuclease-free water and template.
[0059] CMV CIC Sequence 5-3:
[0060] GATAACCAAGCCTGAGGTTATCAATGTCATGAAGCGCCGCATTGAGGAGATCTGCATGAAGGTCTTTGCCCAGTACATTCT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com