Small-molecule inhibitor targeting PD-L1 gene and application of small-molecule inhibitor
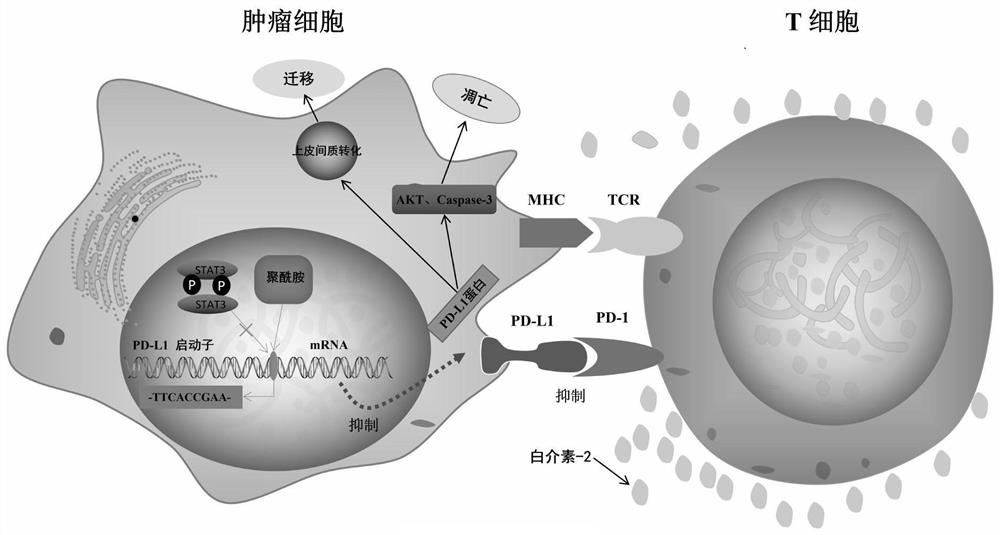
A small molecule inhibitor, PD-L1 technology, applied in the direction of anti-tumor drugs, drug combinations, etc., can solve the problems of less than 30% overall response rate, obvious differential response of patients, and difficult transportation, so as to inhibit tumor immune escape, Inhibit cell migration and enhance the killing effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
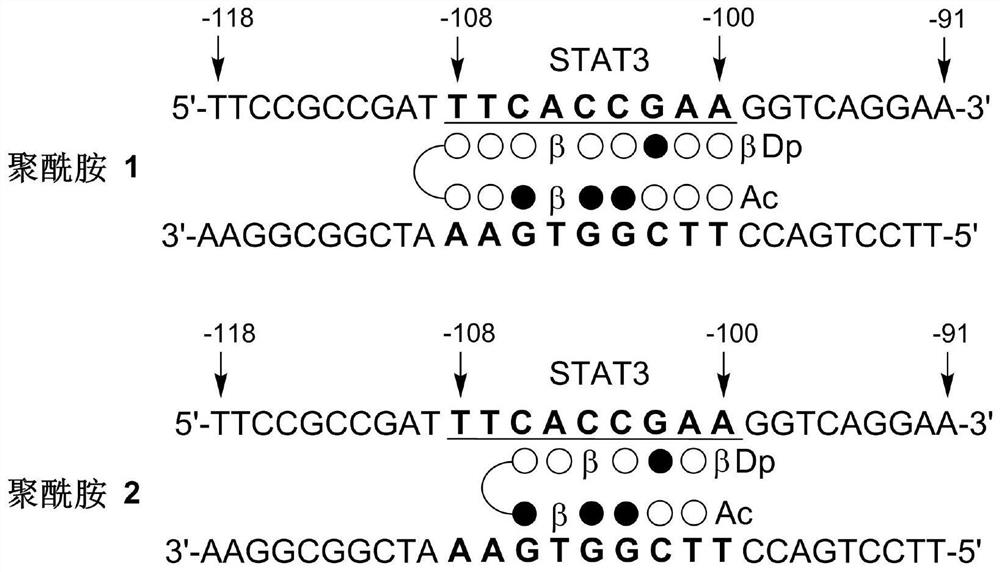
[0032] Embodiment 1: the preparation of polyamide 1
[0033] The abbreviations of the starting materials and reagents used are listed in Table 1.
[0034] Table 1: Reagents and their abbreviations used in solid-phase synthesis reactions
[0035]
[0036] The synthesis steps are as follows:
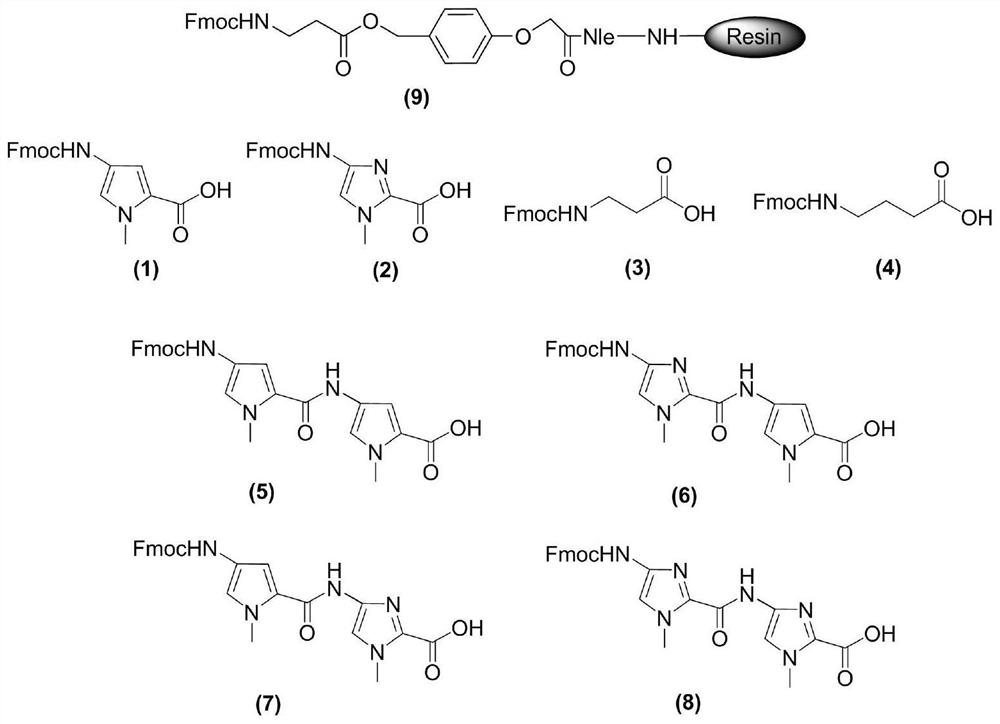
[0037] 1) Activation: Weigh image 3Fmoc protection β-alanine-Clear resin (SPS-1, 1.00g, 0.4mmol, Peptides International) as shown in formula (9) joins in the solid-phase reaction tube of solid-phase reaction device, logical nitrogen protection, again Add 5mL of DMF to fully bubble nitrogen for 30min to activate the resin;
[0038] 2) Deprotection: first prepare 3 mL of 20% (v / v) piperidine / DMF (both piperidine and DMF are treated anhydrous solvents), and add it to the solid-phase reaction tube of step 1) under nitrogen protection, Fully bubbling reaction for 15 minutes, remove the amino protecting group on β-alanine, remove the solvent in the reaction tube, rinse twice with 3mL anh...
Embodiment 2
[0070] Embodiment 2: the preparation of polyamide 2
[0071] The synthesis steps are similar to the synthesis of polyamide 1, and the specific steps are as follows:
[0072] 1) Activation: Weigh image 3 Fmoc protection β-alanine-Clear resin (SPS-1, 1.00g, 0.4mmol, Peptides International) as shown in formula (9) joins in the solid-phase reaction tube of solid-phase reaction device, logical nitrogen protection, again Add 5mL of DMF to fully bubble nitrogen for 30min to activate the resin;
[0073] 2) Deprotection: first prepare 3 mL of 20% (v / v) piperidine / DMF (both piperidine and DMF are treated anhydrous solvents), and add it to the solid-phase reaction tube of step 1) under nitrogen protection, Fully bubbling reaction for 15 minutes, remove the amino protecting group on β-alanine, remove the solvent in the reaction tube, rinse twice with 3mL anhydrous dichloromethane, rinse once with 2mL anhydrous DMF, each time Drain the solvent after rinsing;
[0074] 3) Coupling: Weig...
Embodiment 3
[0097] Embodiment 3: Preparation of mismatched polyamide
[0098] The synthesis steps are similar to the synthesis of polyamide 1, and the specific steps are as follows:
[0099] 1) Activation: Weigh image 3 Fmoc protection β-alanine-Clear resin (SPS-1, 1.00g, 0.4mmol, Peptides International) as shown in formula (9) joins in the solid-phase reaction tube of solid-phase reaction device, logical nitrogen protection, again Add 5mL of DMF to fully bubble nitrogen for 30min to activate the resin;
[0100] 2) Deprotection: first prepare 3 mL of 20% (v / v) piperidine / DMF (both piperidine and DMF are treated anhydrous solvents), and add it to the solid-phase reaction tube of step 1) under nitrogen protection, Fully bubbling reaction for 15 minutes, remove the amino protecting group on β-alanine, remove the solvent in the reaction tube, rinse twice with 3mL anhydrous dichloromethane, rinse once with 2mL anhydrous DMF, each time Drain the solvent after rinsing;
[0101] 3) Coupling:...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com