Non-homology, multi-long-fragment, high-efficiency and zero-background assembly method
A technology of homology and long fragments, applied in the field of bioengineering, can solve the problems of no homology of gene fragments, inability to link 6 fragments into a circle, and many steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
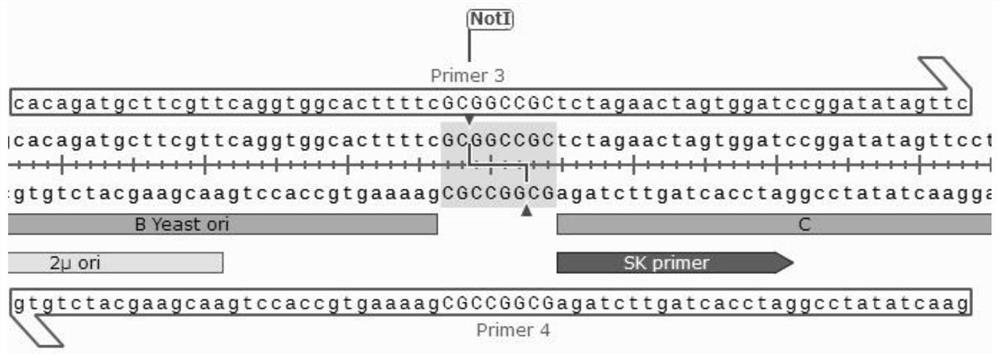
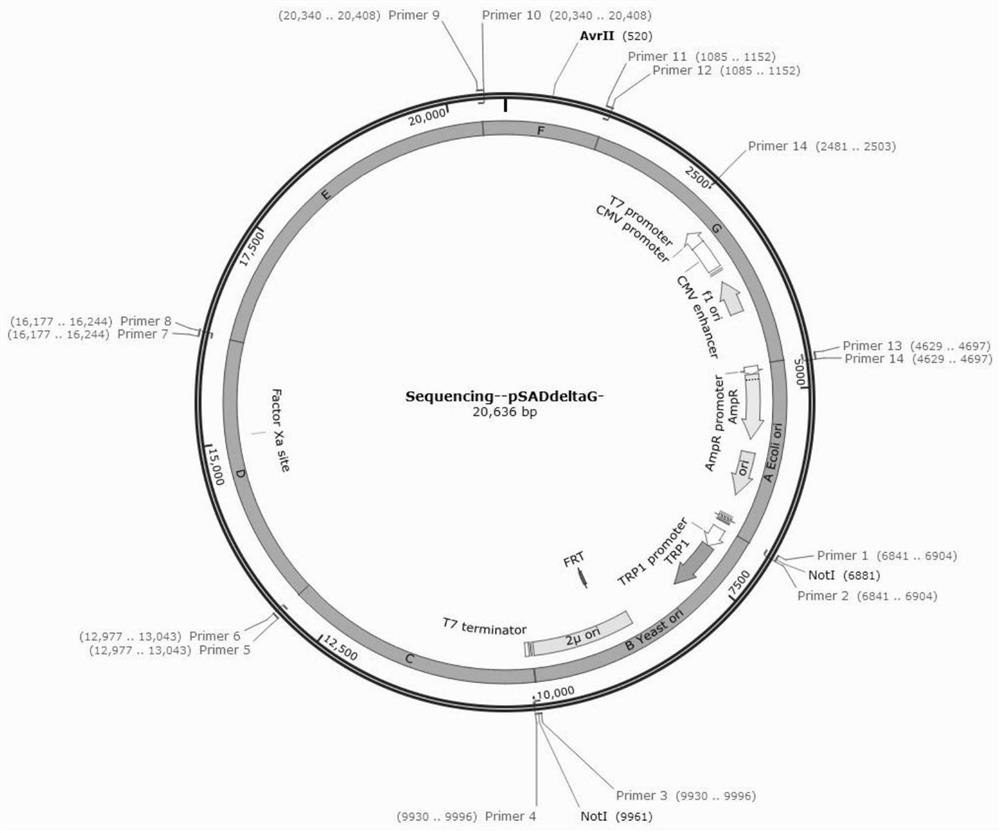
[0028] see Figure 1-3 As shown, no homology, multiple long fragments, efficient zero-background group method, including the following steps:
[0029] The first step is to add a B fragment Yesatori at the only place of the restriction endonuclease NotI site of the entire circularization plasmid, and at the same time ensure that there are exactly two NotI restriction sites on the left and right sides of the B fragment, so that the subsequent NotI single-cut self-ligation Remove the B fragment;
[0030] The second step, design two Primer 1 / 2 primers between fragments A and B without homology, design two Primer 3 / 4 primers between fragments B and C without homology, and use the primers as two fragments The bridges connected between them, and the design of other fragments are analogized in turn;
[0031] The third step is to combine all the fragments with a total of 7 ABCDEFGs. These fragments can be obtained by PCR amplification or plasmid digestion, and a total of 7 pairs of p...
Embodiment 2
[0042] No homology, multiple long fragments, high-efficiency zero-background group method, including the following steps:
[0043] The first step is to add a B fragment Yesatori at the only place of the restriction endonuclease NotI site of the entire circularization plasmid, and at the same time ensure that there are exactly two NotI restriction sites on the left and right sides of the B fragment, so that the subsequent NotI single-cut self-ligation Remove the B fragment;
[0044] The second step, design two Primer 1 / 2 primers between fragments A and B without homology, design two Primer 3 / 4 primers between fragments B and C without homology, and use the primers as two fragments The bridges connected between them, and the design of other fragments are analogized in turn;
[0045] The third step is to combine all the fragments with a total of 7 ABCDEFGs. These fragments can be obtained by PCR amplification or plasmid digestion, and a total of 7 pairs of primer1-14 with a pair...
Embodiment 3
[0056] No homology, multiple long fragments, high-efficiency zero-background group method, including the following steps:
[0057] The first step is to add a B fragment Yesatori at the only place of the restriction endonuclease NotI site of the entire circularization plasmid, and at the same time ensure that there are exactly two NotI restriction sites on the left and right sides of the B fragment, so that the subsequent NotI single-cut self-ligation Remove the B fragment;
[0058] The second step, design two Primer 1 / 2 primers between fragments A and B without homology, design two Primer 3 / 4 primers between fragments B and C without homology, and use the primers as two fragments The bridges connected between them, and the design of other fragments are analogized in turn;
[0059] The third step is to combine all the fragments with a total of 7 ABCDEFGs. These fragments can be obtained by PCR amplification or plasmid digestion, and a total of 7 pairs of primer1-14 with a pair...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com