A paper-based detection method for exosomes
A detection method, exosome technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., to achieve the effect of low requirements, enhanced visual signal, and easy detection and operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0063] Example 1 Establishment of a paper-based detection method for exosomes
[0064] 1. Experimental materials
[0065] 20×SSC, Tris-HCl, sodium phosphate dodecahydrate, bovine serum albumin (BSA), Tween-20, sucrose and TMB chromogenic solution, etc.
[0066] The sequence design is as follows (SEQ ID NO:1-4):
[0067]
[0068] 2. LFB detection and verification of single-stranded DNA targets
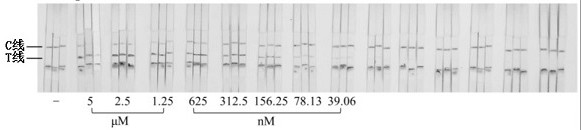
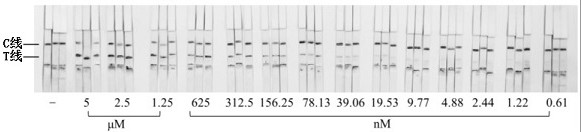
[0069] Dilute the single-stranded DNA target 2-fold concentration gradient, add 50 µL system buffer and 1 µL single-stranded DNA target solution to the sample pool, and drop-coat 1 µL PtAu-NiO on the superposition of the test paper sample pad and the binding pad 2 After the nanozyme signal probe, insert the test paper sample pad into the sample pool, wait for 3 minutes, and observe the result with the naked eye. When both the C line and the T line are black, it is determined to be positive. The color of the T line becomes lighter as the concentration of the ssDNA target decreases,...
Embodiment 2
[0078] Example 2 Practical investigation of detection method
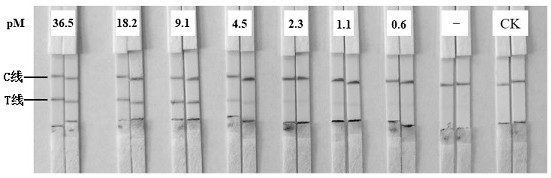
[0079] According to the detection method constructed in Example 1, exosomes extracted from equal volumes of HepG2 and L02 cell supernatants were detected. LFB can effectively distinguish tumor exosomes from non-tumor exosomes ( Figure 6 ).
Embodiment 3
[0080] Embodiment 3 Amplification effect verification of detection method
[0081] After conversion and amplification of exosomes, the gray pixel area of each T line in the LFB detection result was substituted into the gray pixel area of the T line obtained by LFB detection after 2-fold gradient dilution of the single-stranded DNA target, and the standard curve fitting was obtained. Formula: y =14.829e 0.0005x In , the concentration of the single-stranded DNA target after exosome conversion and amplification was obtained. The exosome concentration was converted to the concentration of the substance and compared with the single-stranded DNA target. The molecular magnification of exosomes by this method is 1000 times (Table 1).
[0082] Table 1 Molecular magnification of exosomes after target conversion and amplification
[0083]
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com