Homologous protein detection method based on biological protein information network comparison
A protein detection and information network technology, applied in the fields of bioinformatics, informatics, biological systems, etc., can solve the problems of unsatisfactory prediction results of homologous proteins and poor biological function quality.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
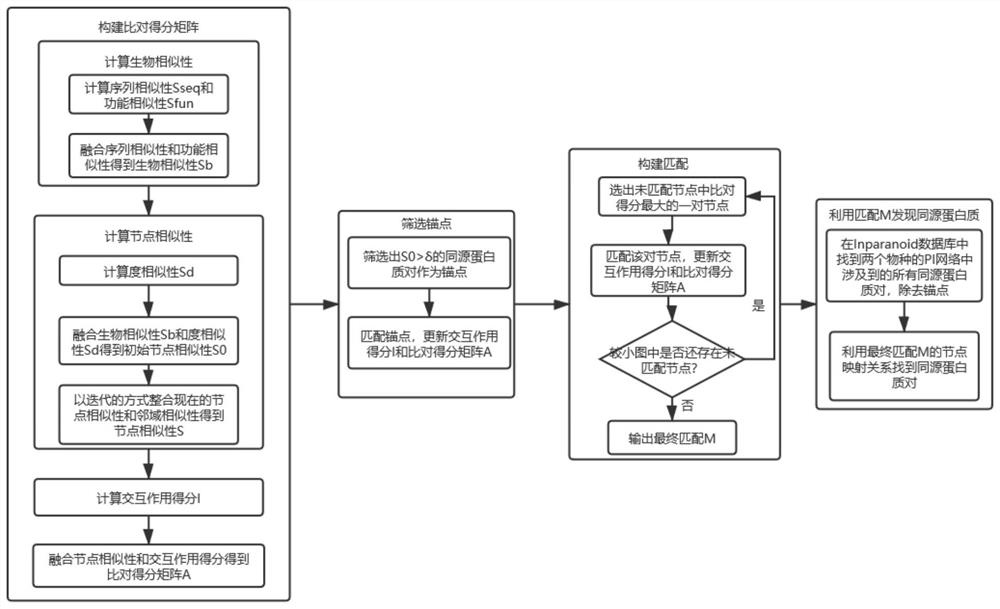
[0029] The present invention is mainly based on a newly proposed biological protein information network comparison algorithm, finds the node matching relationship between protein interaction networks of different species, searches the Inparanoid database according to the matching results, and finds homologous protein pairs between species. Through the present invention, we provide a new method for detecting homologous protein pairs between different species, which is more effective than traditional methods, and compared with the existing PPI network comparison algorithm, this algorithm can better balance Topological quality and biofunctional quality of the matching results.
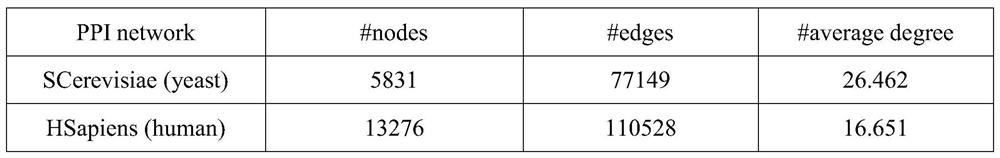
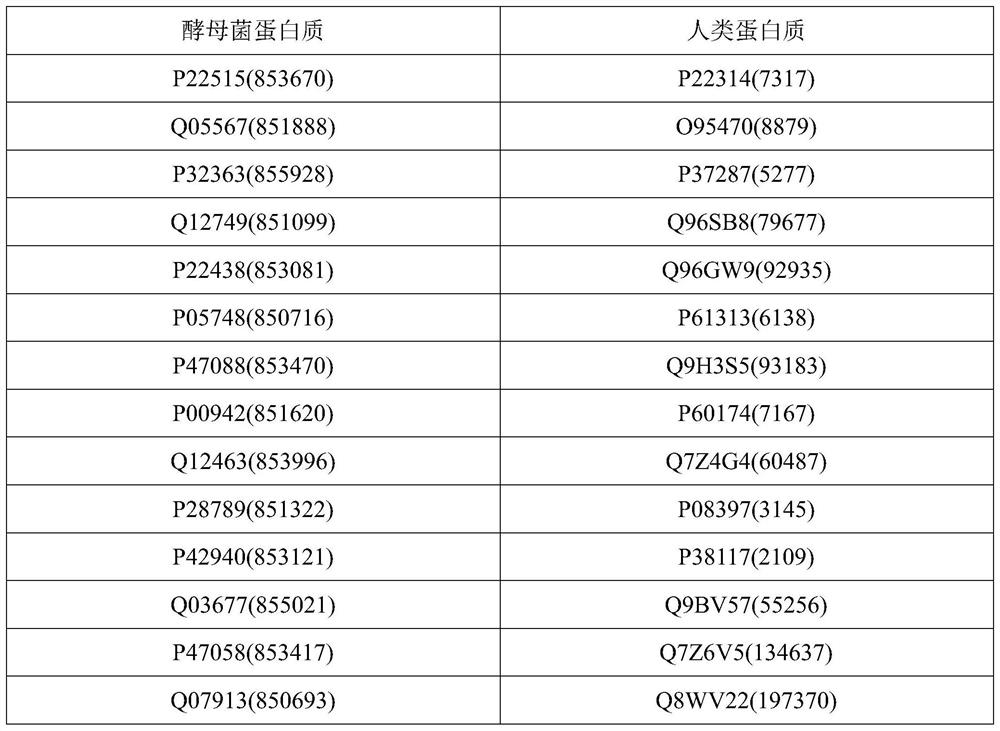
[0030] The method provided by the invention can use computer software technology to realize the process. see figure 1 , the embodiment takes the protein interaction network (PPI network) of yeast (yeast, biological code SCerevisiae) and the protein interaction network (PPI network) of human (human, biolo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com