Promoter of nosema bombycis inducible expression gene BmPUGT2 and application of promoter
A technology of silkworm microparticles and inducible expression, applied in microorganisms, animal cells, glycosyltransferases, etc., to achieve the effect of good application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
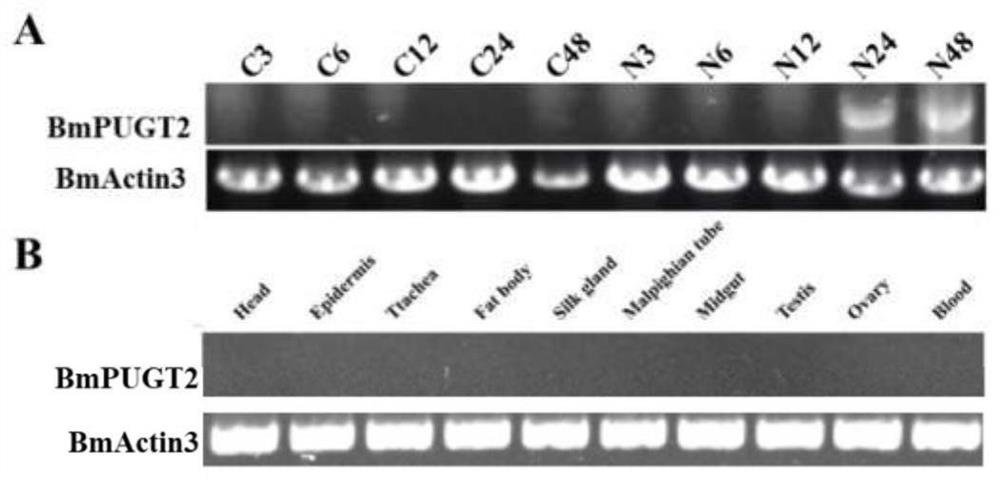
[0021] Silk silkworm micropelic induced Bmpugt2 gene expression
[0022] Depending on the silkworm genomic database Silkdb (https: / / silkdb.bioinfotoolkits.net / main / species-info / -1) and NCBI (https: / / www.ncbi.nlm.nih.gov), get BMPUGT2 (AK378453.1) Gene's CDS sequence (SEQ ID NO: 1), such as figure 1 As shown, BMPUGT2 gene sequence diagram, the hemorrhage bold ATG represents the translation start site, the TGA indicates the termination codon, the gray frame represents the non-translation zone, and the detection primer of the gene is designed according to the gene sequence of Bmpugt2. Bmpugt2-f and bmpugt2 -R, the silkworm ACTIN 3 gene is innergin, the primer is BMA3-F and BMA3-R.
[0023] Bmpugt2-f: 5'-AtcattcacagTacgtat-3 '(SEQ ID NO: 2)
[0024] Bmpugt2-R: 5'-Gaaacattattattgt-3 (SEQ ID NO: 3)
[0025] BMA3-F: 5'-AtggggCgctcctccaagaacg-3 '(SEQ ID NO: 4)
[0026] BMA3-R: 5'-ctacaggaacaggtggggTGGCGGG-3 '(SEQ ID NO: 5)
[0027] Made of normal silkworm breeding in standard environment...
Embodiment 2
[0029] BMPUGT2 promoter carrier construction
[0030] Depending on the silkworm genomic database (https: / / silkdb.bioinfotoolkits.net / main / species-info / -1), the genomic sequence of the first 1247 bp of the BMPugt2 gene (SEQ ID NO: 6), such as image 3 As shown, the lowercase the 5'utr area of BMPugt2, the upper-write slope bold ATG represents the translation start site, using the website to analyze this genome sequence (https: / / www.fruitfly.org / seq_tools / promoter .html) found that in this sequence contains a typical promoter structure area, the region-specific primer PPUGT2-F-ECORI and PPUGT2-R-BamHi amplification of the sequence, the sequence is as follows:
[0031]
[0032]
[0033] The genomic extract kit (D3396, Omega) extracts the genomic DNA of the silkworm lottery, and uses primer PPUGT2-F-EcoRi, PPUGT2-R-BAMH and Q5 high-fidelity enzyme (M0493, NEB) to carry out PCR Amplification, its amplification conditions were: 98 ° C pre-degeneration 30s; 98 ° C denaturation 10s,...
Embodiment 3
[0036] BMPUGT2 promoter function verification
[0037] The PSL [PPUGT2-MCHERRY-SV40] expression vector constructed in Example 2 was transfected into the BMN-SWU1 cell line according to the plasmid: liposome = 1: 2, Ug: ul, and added silkworm microparticles, and silkworm. The ratio ratio of the fine particles and cells was 10: 1 to add PBS for control, and the red fluorescence was detected by fluorescence microscopy after 72 h. Such as Figure 4 As shown, in the cells transfected with PSL [PPUGT2-MCHERRY-SV40], when the silkworm micropel is added, the promoter PPUGT2 is activated and induces the expression of the downstream MCHERRY gene, and the cells are red-fluorescent, and the control group PBS is No red fluorescence, thereby explaining that the promoter PPUGT2 is the purpose of activating activation, achieving regulation, which can be activated by silkworm fine particles.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com