Primers and methods for identifying Rhododendron xing'an and Rhododendron japonica
A technique for rhododendron and its products, which is applied in the field of primers for identifying Rhododendron xing'an and Rhododendron chinensis, can solve the problems of identification and the inability to separate the two species
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
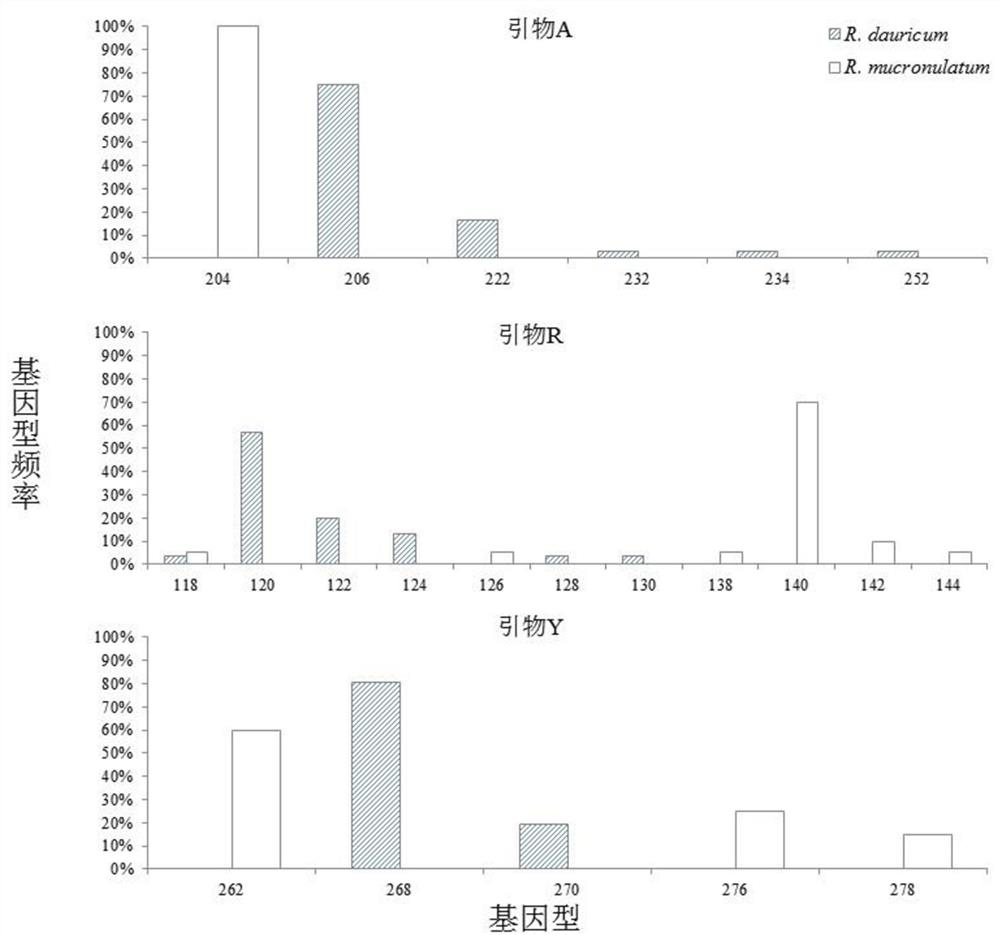
[0027] Example 1 Design of primers
[0028] Based on the previously sequenced SLAF data (PRJNA589346) of Rhododendron xing'an and Rhododendron japonica, and based on the reference genome Rhododendron lantana (R. delavayi), all reads of all samples were aligned and clustered using BLAT software to obtain the SLAF tag sequence. Afterwards, use MISA-web (http: / / misaweb.ipk-gatersleben.de / ) to scan these polymorphic SLAF sequences for microsatellite sites, and set the repeating unit as a microsatellite site of two bases The minimum number of repeats is 6, and the minimum repeats of three, four, five and six bases for the rest of the repeating units are all set to 5 times. Two microsatellite loci are regarded as a microsatellite locus if the distance between them is less than 100bp.
[0029] At the same time, the software SAMtools and GATK were used to identify the insertion / deletion sites of the population, and the parameters of the software were the default parameters. The obta...
Embodiment 2
[0034] Embodiment 2SSR-PCR and product detection
[0035] (1) Use the 4 × CTAB method to extract the whole genome DNA, and use 1% agarose gel for detection to ensure that the concentration reaches 20ng / ul or more, and the DNA that passes the test is stored at -20 ℃;
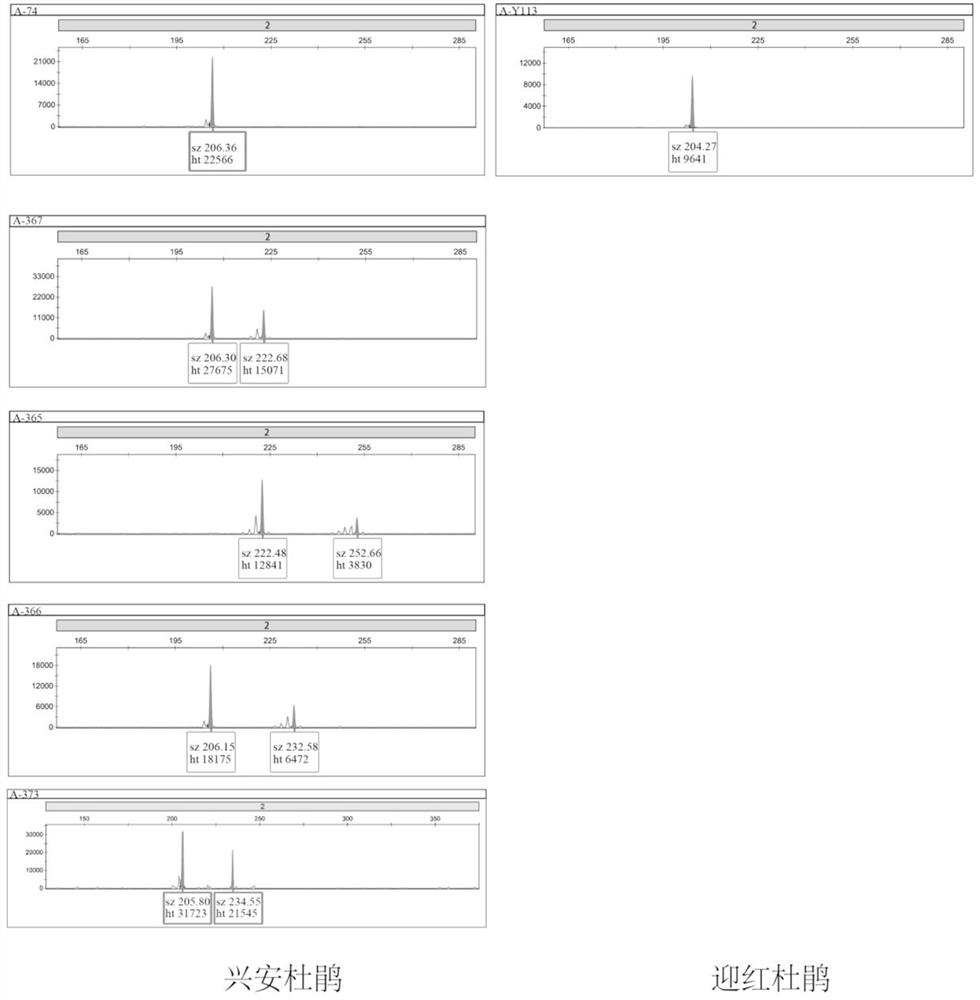
[0036] (2) Use 20 μL fluorescent PCR system for population PCR amplification: 50 ng of the above genomic DNA, 1 × PCRbuffer (plus Mg2+), 0.2 mMdNTP and 0.5 μM each of the forward and reverse primers shown in Table 1, and the forward primers use different Fluorescent markers (FAM, TAMRA, or HEX) (Invitrogen) and 1U enzyme (Takara). PCR amplification was performed using the following program: pre-denaturation at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 10 s in each cycle , anneal for 30 s (the annealing temperature of primers A and R is 51°C, and the annealing temperature of primer Y is 45°C) and extend at 72°C for 30 s, and finally extend at 72°C for 8 min.
[0037] (3) The amplified prod...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com