Development method of strawberry functional gene linked SSR marker
A technology for functional genes and strawberries, which is applied in the development of strawberry functional gene-linked SSR markers, can solve the problems that the development of SSR markers has not yet been reported, and achieves the effects of high polymorphism and good repeatability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
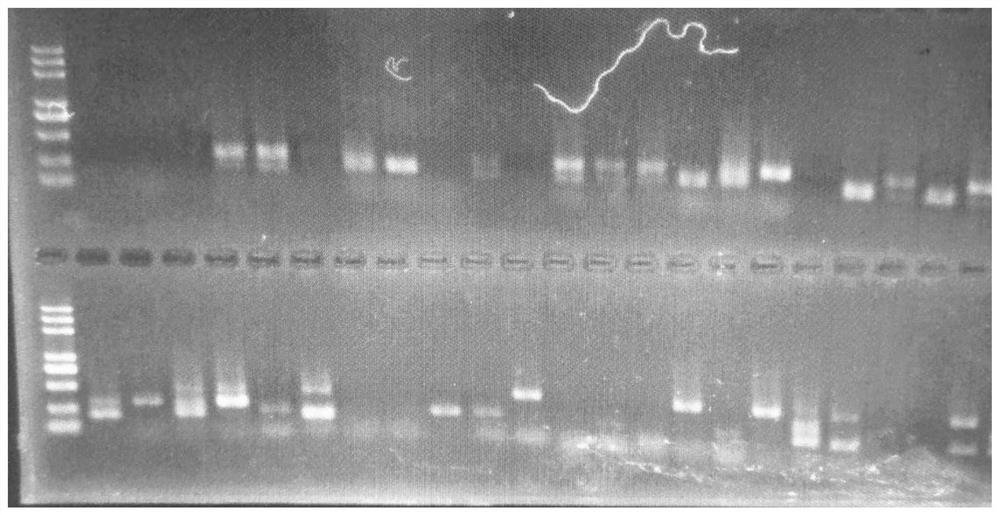
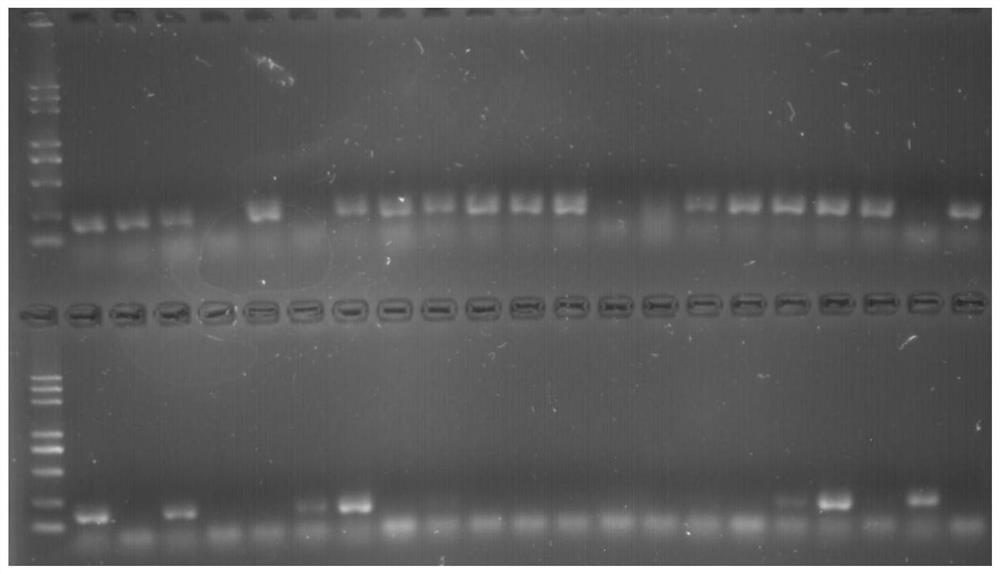
[0026] like Figure 1-4 As shown, combined with the published strawberry genome (https: / / www.ncbi.nlm.nih.gov / bioproject / ?term=PRJNA5083890), use the Fastq-Dump program to convert the SRA format data into Fastq format files, and use parallel The cleaning channel cleans up the converted files to control data quality, including Q20 (1% base error rate) cleaning and L40 (length ≥ 40bp) filtering, and removes polyT, polyA sequences and carrier sequences at the 5 and 3 ends. Subsequently, Trinity software was used to assemble and assemble the quality-controlled sequences according to the default parameters, and remove redundant sequences to obtain unigene. Use the MISA software (http: / / pgrc.ipk-gatersleben.de / misa / ) to identify the SSR site of the unigene sequence. The identification condition is that the single nucleotide repeat is not less than 10 times, and the dinucleotide repeat is not less than low At 8 times, trinucleotide repeats are not less than 7 times, tetranucleotide ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com