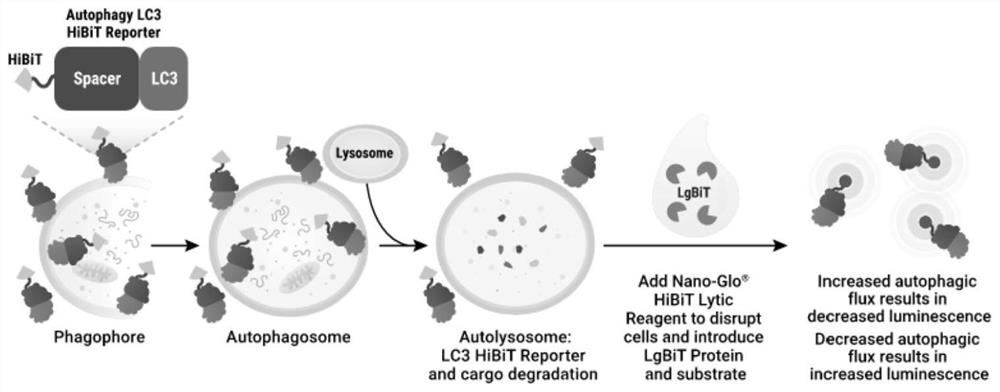
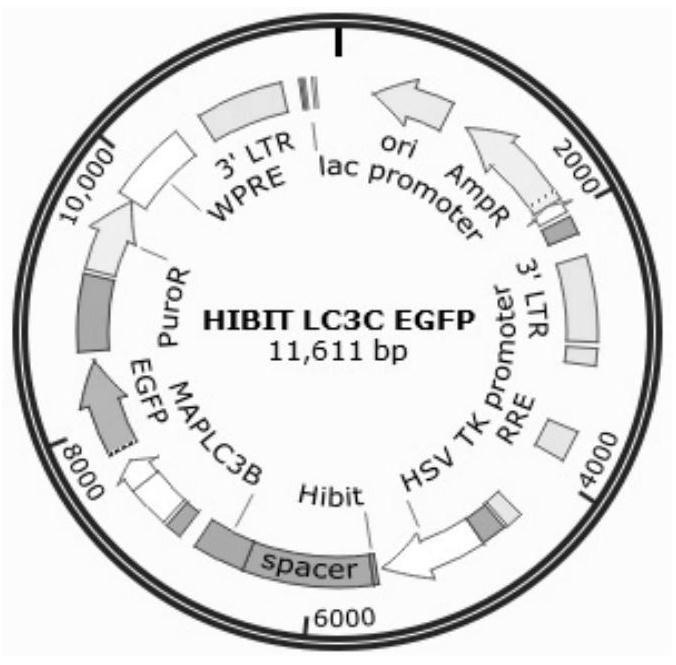
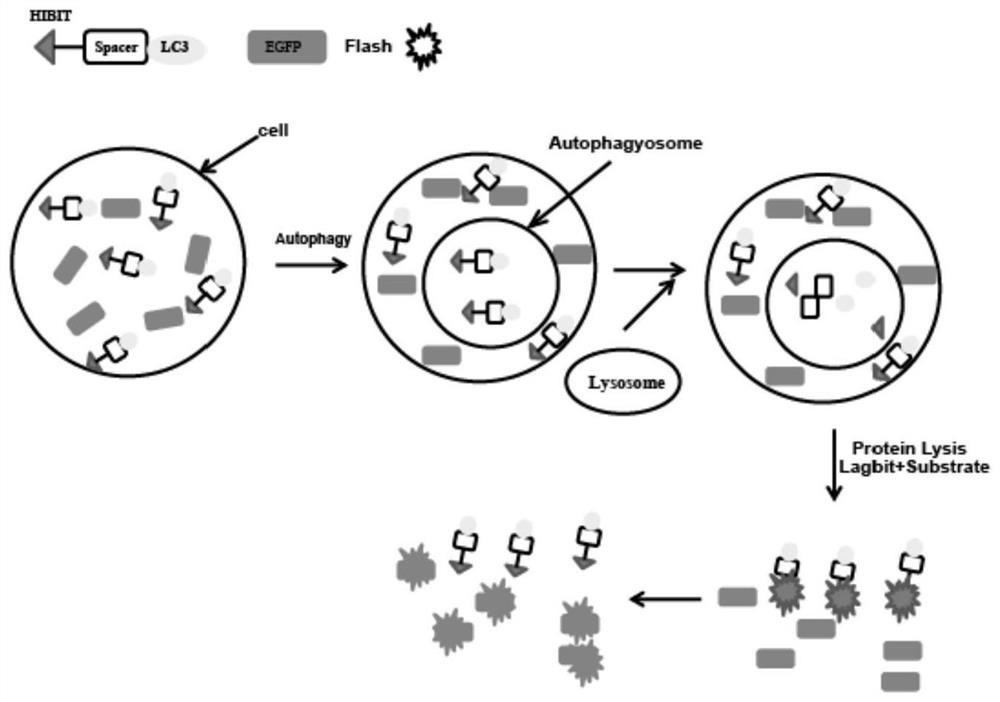
Method for accurately and quantitatively detecting autophagy flux
A quantitative detection and autophagy technology, which is applied in botany equipment and methods, biochemical equipment and methods, viruses/bacteriophages, etc., can solve problems such as the inability to rule out systematic errors in reading values between groups and limited application range, and achieve expansion Scope of application and detection accuracy, save time and cost, and promote the effect of research and development
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] 1. Construct EGFP / LC3 HIBIT co-expression vector. pattern such as figure 2 shown.
[0027] Specific implementation steps: According to the method of seamless cloning, design independent EGFP fragments containing promoters and LC3HIBIT linearization primers respectively, and use KOD enzyme to amplify. System: 10×Buffer for KOD-Plus-5μL, 2mMdNTPs 5μL, 25mM MgSO4 2μL, Primer F 1.5μL, Primer R 1.5μL, Template DNA XμL (Plasmid DNA1-50ng / 50μL), KOD-Plus-1μL, water up to 50μL . Program: 94°C, 2min; 94°C, 15s, Tm-[5-10]°C, 30sec, 35cycle; 68°C, 1min. / kb. Electrophoresis and recovery of target fragments. Then prepare a seamless cloning reaction system: fragment molar ratio 1:1, 5×TEDA 4 μL, water up to 20 μL. Program: 30°C, 40min. Finally, the correct clones were screened by transformation sequencing.
[0028] 2. Transiently transfect or package lentivirus to infect cells, so that the cells co-express EGFP and LC3-HIBIT proteins. Specific implementation steps: transient...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com