Gene sets, scoring models and their applications for assessing the tumor microenvironment
A tumor microenvironment and gene set technology, applied in the field of bioinformatics, can solve the problems of incomplete immunotherapy mechanism, low response rate, poor stability, etc., achieving significant prediction effect, saving medical resources, and better efficacy Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Example 1 Multi-gene scoring model of tumor microenvironment of the present invention and its construction method
[0063] A method for constructing a multi-gene scoring model for evaluating tumor microenvironment includes the following steps:
[0064] (1) standardized data of Affymetrix chip of 300 patients with gastric cancer; Said data originated from GSE62254 (obtained from NCBI public database);
[0065] (2) evaluating the components of immune cells in tumor microenvironment by using CIBERSORT and MCP-counter algorithm to obtain the components evaluation results of 23 immune cells;
[0066] (3) Unsupervised clustering of immune cells in tumor microenvironment was carried out by consensuclusterplus2 to obtain different tumor microenvironment types;
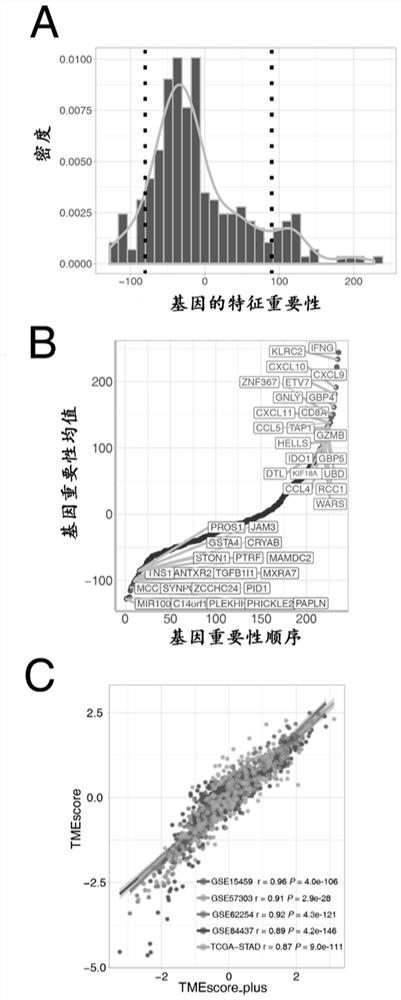
[0067] (4) Through pairwise difference analysis and random forest dimensionality reduction, 244 gene sets that can evaluate tumor microenvironment scores were finally determined;
[0068] (5) In order to further realize the...
Embodiment 3
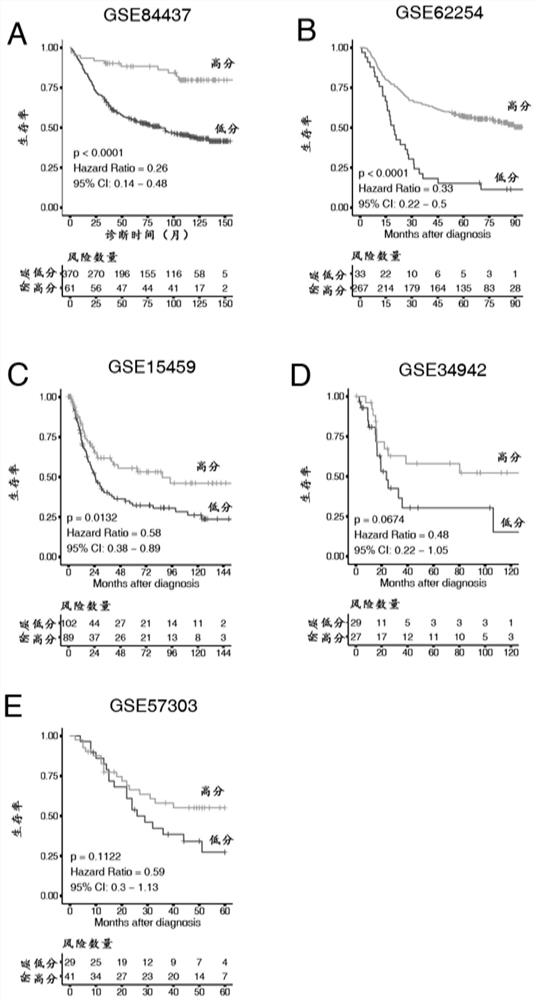
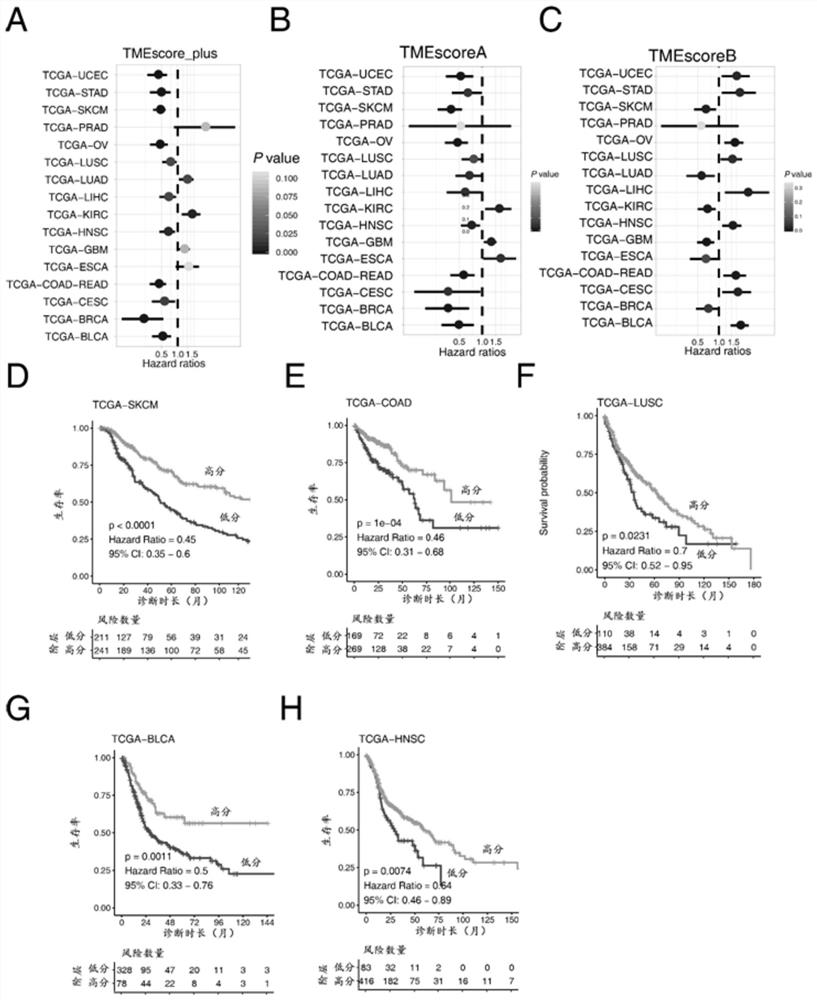
[0090] Example 3 Evaluation of the Efficacy of Tumor Microenvironment Scoring Model in Predicting the Prognosis of Patients
[0091] The above tumor microenvironment scoring system is applied to a number of gastric cancer patient queues with survival data, and the prognostic value of tumor microenvironment is analyzed. The steps are as follows:
[0092] ① Get the transcriptome chip data of gastric cancer patients from the website (https: / / www.ncbi.nlm.nih.gov / geo / ) (GSE84437, GSE15459, GSE34942, GSE57303, GSE6254), We downloaded the patient's original chip data and standardized and corrected the data by affy (Ref. 14: L. Gautier, L. Cope, B. M. Bolstad, R. A. Irizarry, AFFY-analysis of affymetrix gene chip data at the probe level. Bioinformatics 20, 307-315 (2004
[0093] ② The data of 44 tumor microenvironment genes in the gene expression matrix were extracted, and after Z-score transformation (standardized as data with mean value of 0 and standard deviation of 1), the tumor immu...
Embodiment 4
[0097] Example 4 Evaluation of Efficacy of Tumor Microenvironment Scoring Model in Predicting Immunotherapy Response of Tumor Patients
[0098] To verify the efficacy of TMEscore_plus in predicting immunotherapy, the steps are as follows:
[0099] ① Download the second-generation RNAseq sequencing data of PRJEB25780 (https: / / www.ebi.ac.uk / ENA / browser / view / PRJEB25780), and standardize the data of samples by TPM;
[0100] ② The data of 44 tumor microenvironment genes in gene expression matrix are extracted, and the sample data are standardized, so that the mean value of each gene is 0 and the standard deviation is 1, and the expression matrix is assigned to eset objects in R;
[0101] ③ Install the TMEscore R package, and use the following code to calculate the tumor microenvironment score:
[0102] # install TMEscore package
[0103] devtools::install_github("DongqiangZeng0808 / TMEscore")
[0104] # Load R package
[0105] library('TMEscore')
[0106] # Enter the gene expression mat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com