Marker, reagent or kit for detecting whether COVID-19 disease is cured
A coronavirus, a new type of technology, applied in the field of reagents or kits, markers to detect whether the new coronavirus disease is cured, can solve the problem of no biomarkers for the occurrence, development and outcome of coronavirus disease
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
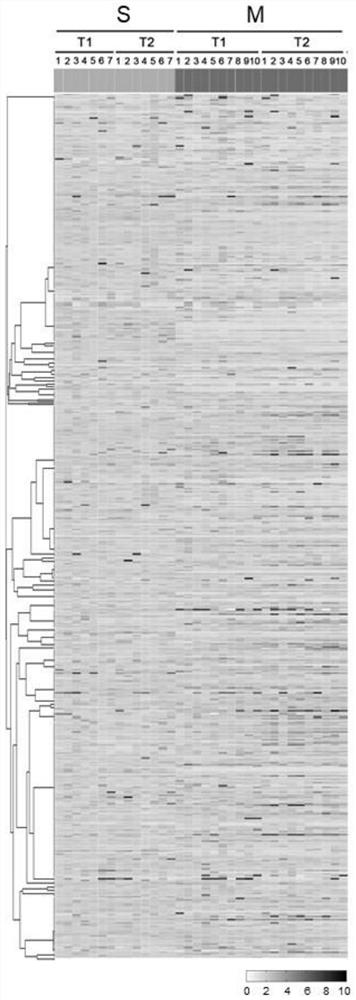
[0033] Example 1: Mass Spectrometry Analysis of Plasma Samples from Patients with New Coronary Pneumonia
[0034] 1. Experimental materials
[0035] Plasma samples from COVID-19 patients, including whole blood collected from 10 mild patients (M1T1-M10T1, referred to as MT1), 7 severe patients (S1T1-S7T1, referred to as ST1) and 10 mild patients Whole blood collected before discharge (M1T2-M10T2, referred to as MT2) and whole blood collected from 7 critically ill patients before discharge during hospitalization (S1T2-S7T2, referred to as ST2) totaled 34 whole blood samples from a hospital.
[0036] According to the "Diagnosis and Treatment Plan for Pneumonia Infected by Novel Coronavirus (Trial Version 6)" jointly issued by the General Office of the National Health Commission and the Office of the State Administration of Traditional Chinese Medicine, the clinical classification is as follows: mild, mild clinical symptoms, no pneumonia manifestations in imaging ;Severe, meeting...
Embodiment 2
[0044] Example 2: Machine learning screening for markers of cure in COVID-19 patients
[0045] 1. Experimental materials
[0046] Mass spectrometry data of 34 plasma samples (source Example 1), Python 3.7 (https: / / www.anaconda.com / ), Scikit learn 0.22.1 (https: / / scikit-learn.org / stable / ). The source code of this experiment is https: / / github.com / Ning-310 / POC-19.
[0047] 2. Experimental process
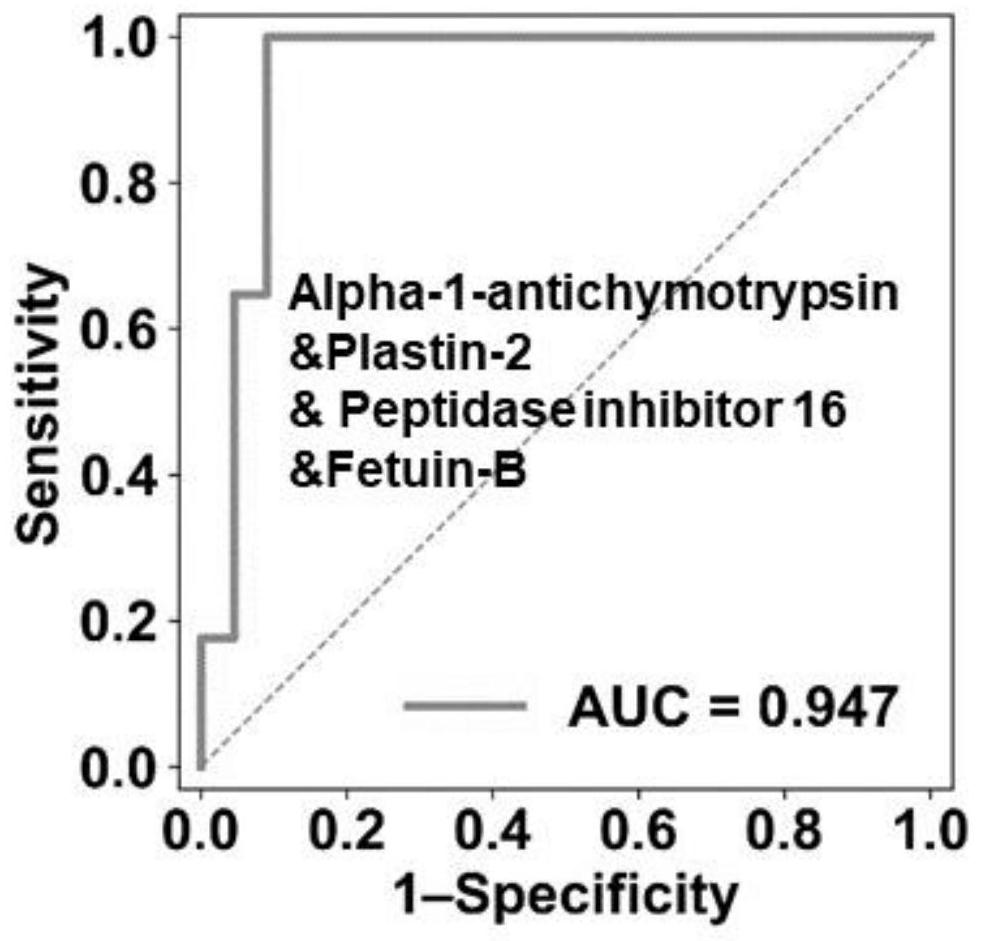
[0048] (1) Screen for differential proteins, screen for protein fold change (FC) absolute value greater than 0.8 in ST1+MT1 group and ST2+MT2 group, and two-tailed unpaired Welch T test is less than 0.01 (|log2(FC)| > 0.8, unpaired two-sided Welch'st-test, P < 0.01).
[0049] (2) Randomly select no more than 5 proteins from the differential proteins to form a potential optimal marker combination (OBC). The initial weight value of each protein is set to 1, and 1000 OBC candidates are set.
[0050] (3) For each candidate OBC, we randomly generate a training dataset and a testing data...
Embodiment 3
[0058] Example 3: Validation of OBC Predictor Accuracy
[0059] 1. Experimental materials
[0060] Mass spectrometry data of 34 plasma samples, Python 3.7 (https: / / www.anaconda.com / ).
[0061] 2. Experimental process
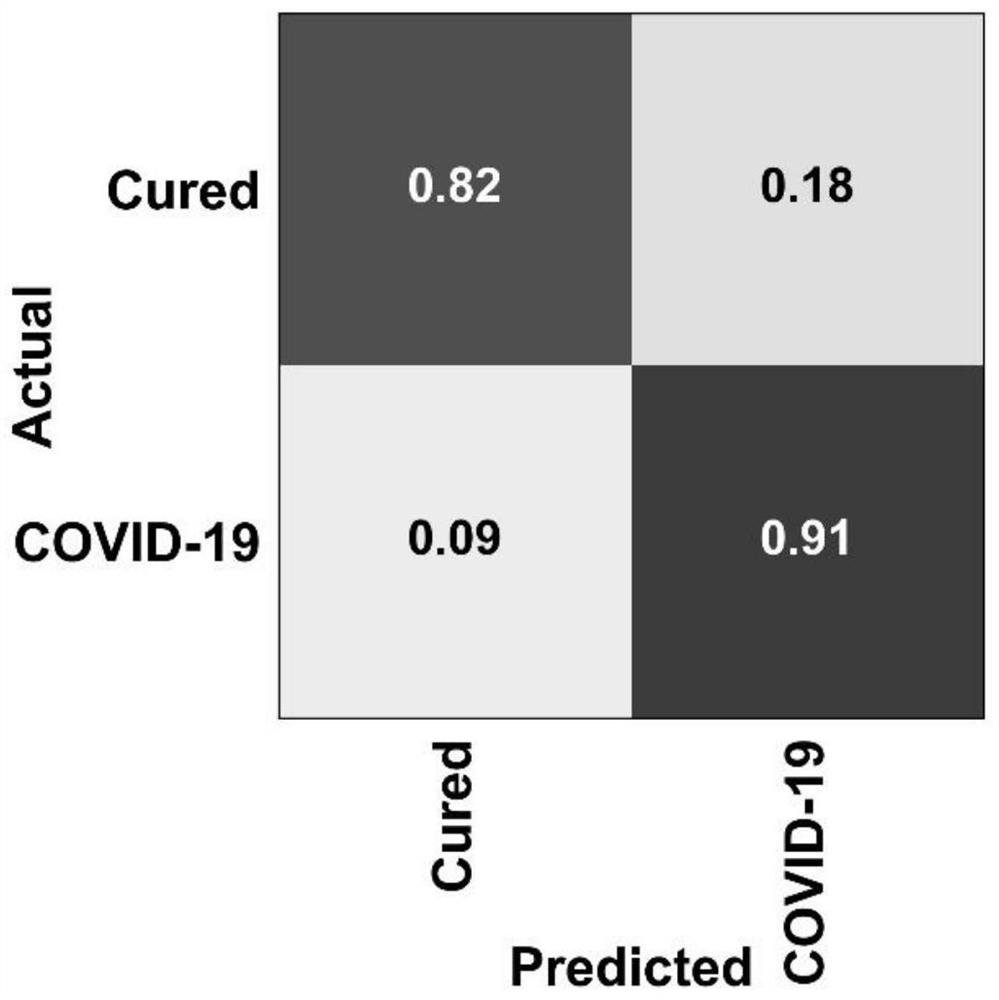
[0062] (1) Differentiate each protein in OBC identified as true positive (TruePositive, TP), true negative (True Negative, TN), false positive (False Positive, FP) and false negative of the possibility of COVID-19 patients and cured patients (FalseNegative, FN) ratio for chaotic logic analysis.
[0063] (2) Use Scikit learn 0.22.1 software to draw the chaos matrix.
[0064] The result is as image 3 As shown, the OBC combination predicts the true positive rate of 82% and the false positive rate of 18.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com