Spatial transcriptome constructing method
A technology of spatial transcription and construction methods, applied in the fields of biochemical equipment and methods, genomics, proteomics, etc., can solve the problems of complex experimental operation, achieve a wide range of applications, speed up analysis, reduce costs and operational complexity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
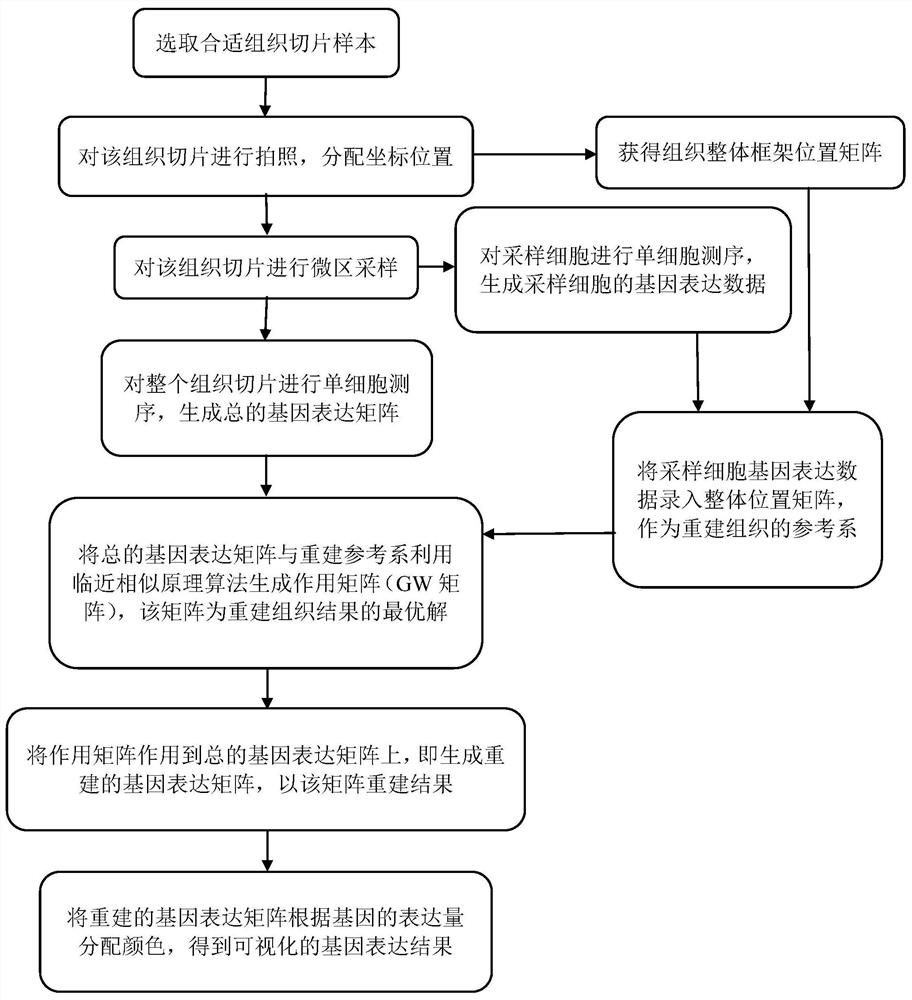
[0042] Construction of a spatial location map of adult cardiac tissue.
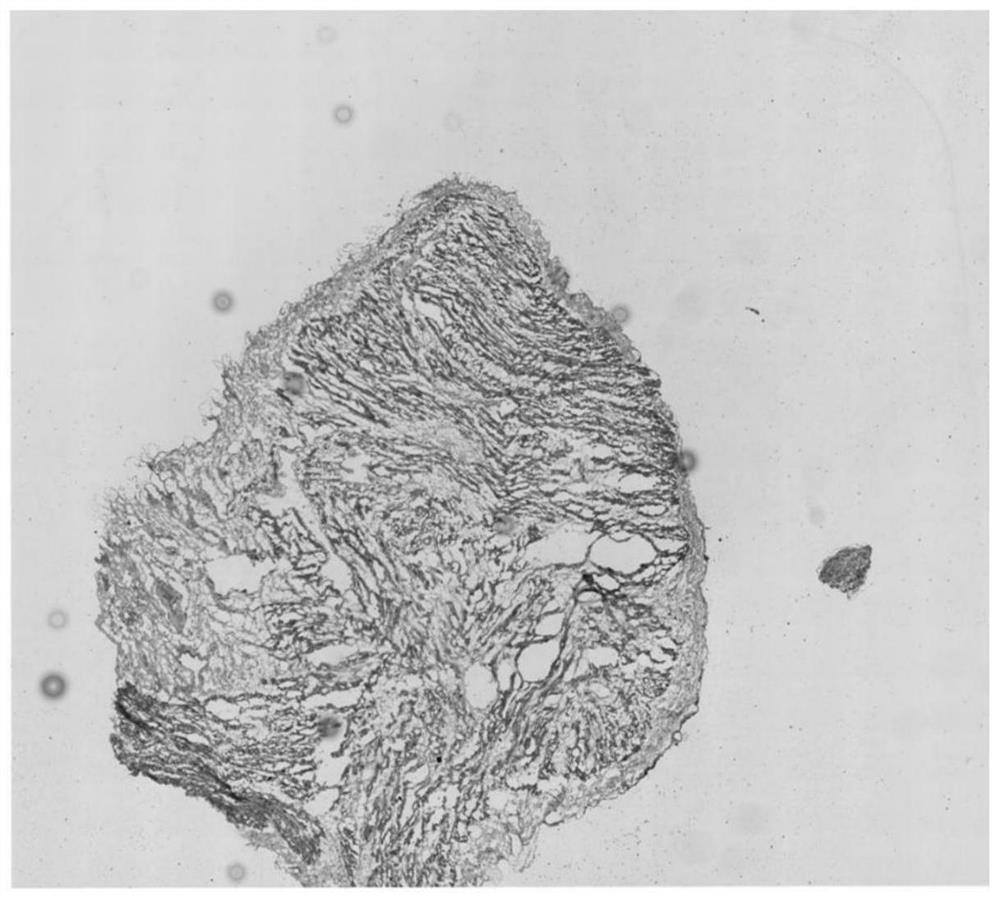
[0043] 1. Obtain images of adult heart tissue slices
[0044] Determine the tissue samples, select tissue samples with clear outlines to take pictures, and generate tissue slice pictures (see figure 2 ).
[0045] 2. Microplot Sampling Cells
[0046] Single cells are collected from different regions in tissue sections, and single-cell RNA sequencing (ie, single-cell cytoplasmic mRNA sequencing) is performed on the sampled cells, and the gene expression data and location information of the sampling points are recorded.
[0047] 3. Determination of single-cell expression data in adult heart tissue
[0048]Using single-cell RNA sequencing technology (i.e., single-cell cytoplasmic mRNA sequencing) to measure the transcriptome data of all cells in adult heart tissue samples, the raw data are compared to obtain the expression levels of all genes in each cell.
[0049] 4. Data preprocessing
[0050] Express...
Embodiment 2
[0059] Construction of spatial location map of mouse hippocampus.
[0060] 1. Obtain pictures of mouse hippocampal tissue slices
[0061] Determine tissue samples, select tissue samples with clear outlines and take pictures, and generate tissue slice pictures.
[0062] 2. Microplot Sampling Cells
[0063] Cells were collected from different areas in the tissue section, and single-nuclear RNA sequencing was performed on the sampled cells, and the nuclear gene expression data and location information of the sampling points were recorded.
[0064] 3. Determination of single-nucleus gene expression data of mouse hippocampus tissue
[0065] Single-nucleus RNA sequencing technology was used to measure the single-cell transcriptome data of all cells in the mouse hippocampus tissue sample, and the original data were compared to obtain the expression of all genes in each nucleus.
[0066] 4. Data preprocessing
[0067] Expression Data Extract raw gene expression data, import the fi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com