A method for reducing concatenation of double-stranded DNA fragments in CRISPR-Cas9 gene editing and its application
A technology of gene editing and fragmentation, which is applied in the field of genetic engineering, can solve problems affecting technical accuracy, methods of reducing concatenation, gene targeting failure, etc., and achieve the effect of eliminating DNA concatenation, reducing concatenation, and reducing concatenation rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
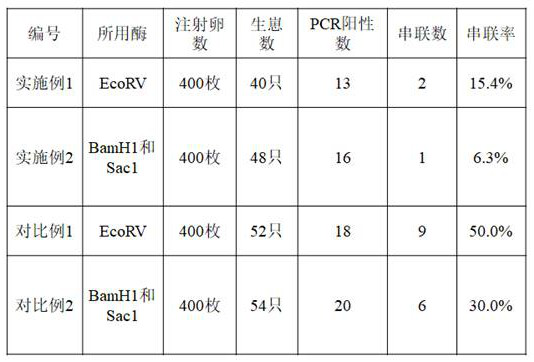
[0051] In this example, a method for reducing the concatenation of double-stranded DNA fragments in CRISPR-Cas9 gene editing is provided.
[0052] (1) Construction of dsDNA
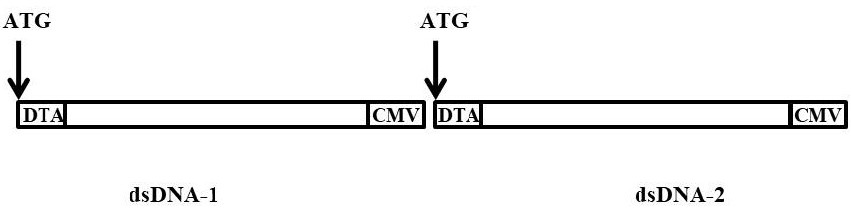
[0053] The schematic diagram of the structure of the dsDNA constructed in this example is as follows figure 1 shown;
[0054] Put DTA forward at the 5' end of dsDNA, CMV forward at the 3' end of dsDNA, and both ends of the dsDNA fragment contain EcoRV restriction sites;
[0055] Construct the designed sequence on the plasmid (Plasmid) vector, and use EcoRV to digest and purify to obtain dsDNA;
[0056] Among them, the enzyme digestion system is EcoRV 3 μL, plasmid 25 μL (concentration is 2 μg / μL), Buffer smart 10 μL, ddH 2 Make up to 100 μL with O;
[0057] Enzyme digestion process: time 8 h; temperature 37°C;
[0058] After digestion, purify and recover, and dilute the dsDNA to 5 ng / uL with TE buffer.
[0059] (2) Microinjection of mouse fertilized eggs
[0060] Obtain fertilized eggs, prepare fix...
Embodiment 2
[0074] In this example, DTA is placed forward at the 5' end of dsDNA, and CMV is placed forward at the 3' end of dsDNA. At the same time, both ends of the dsDNA fragment contain restriction sites for BamH1 and Sac1, and the designed sequence is constructed on the plasmid The vector was digested with BamH1 and Sac1 and purified to obtain dsDNA.
[0075] The digestion system is: BamH1 3 μL, Sac1 3 μL, plasmid 25 μL (concentration is 2 μg / μL), Buffer smart10 μL, ddH 2 Make up O to 100 μL; enzyme digestion process: time 8 h; temperature 37°C;
[0076] The steps of microinjection of mouse fertilized eggs were the same as in Example 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com