Phthalate ester degrading enzyme gene, its encoded product and preparation method
A technology for phthalic acid and degrading enzymes, which is applied in the field of enzyme genetic engineering and can solve the problems of discovering phthalate degrading enzymes and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Embodiment 1: Cloning and transformation of esterase gene
[0049] 1. The soil sample of the present invention comes from the wild ferulic distribution area of Shihezi Nanshan in the southern margin of Jungar Basin, Xinjiang, the soil 5-10cm below the surface. Weigh 1.25g of soil samples into 5mL EP tubes, add 1.5mL of DNA extraction solution to each tube, mix by inverting, and shake on a shaker at 37°C and 130rpm for 30min. Add 400ul 10% SDS to each tube and mix well, shake in 65°C water bath for 2h every 15min, centrifuge at 10000rpm for 10min, take the supernatant and add 0.7 times the volume of isopropanol, discard the supernatant by centrifugation, add an appropriate amount of 70% ethanol to wash, and then Dissolve with appropriate amount of TE buffer to obtain total DNA.
[0050] 2. Use the DNA Extraction Kit to purify and recover the total DNA and store it at 20°C for later use. The purity and quality of the total DNA were checked by agarose gel electrophores...
Embodiment 2
[0051] Embodiment 2: Amplification and expression of esterase gene
[0052] According to the comparison between the sequencing results and NCBI, PCR primers were designed, and two restriction sites, HindIII and BglII, were introduced into the primers. The primer sequences are as follows:
[0053] est1012-HindIII-F:5'-CCC AAGCTT GGCCTTTGGGTGCGTTATACTGAAG-3' (the underlined part is the HindIII restriction site);
[0054] est1012-Bg1II-R:5'-GGA AGATCT The underlined part of CAGCTTCTTTTCGGCCTTTGCTAGCAG-3' is the BglII restriction site).
[0055] Using the plasmid containing pUC118-est1012 as a template, using Prime STAR TM Max Premix for PCR amplification,
[0056] The system is as follows:
[0057] Table 1
[0058]
[0059] The amplification conditions are as follows:
[0060] Table 2
[0061]
[0062]
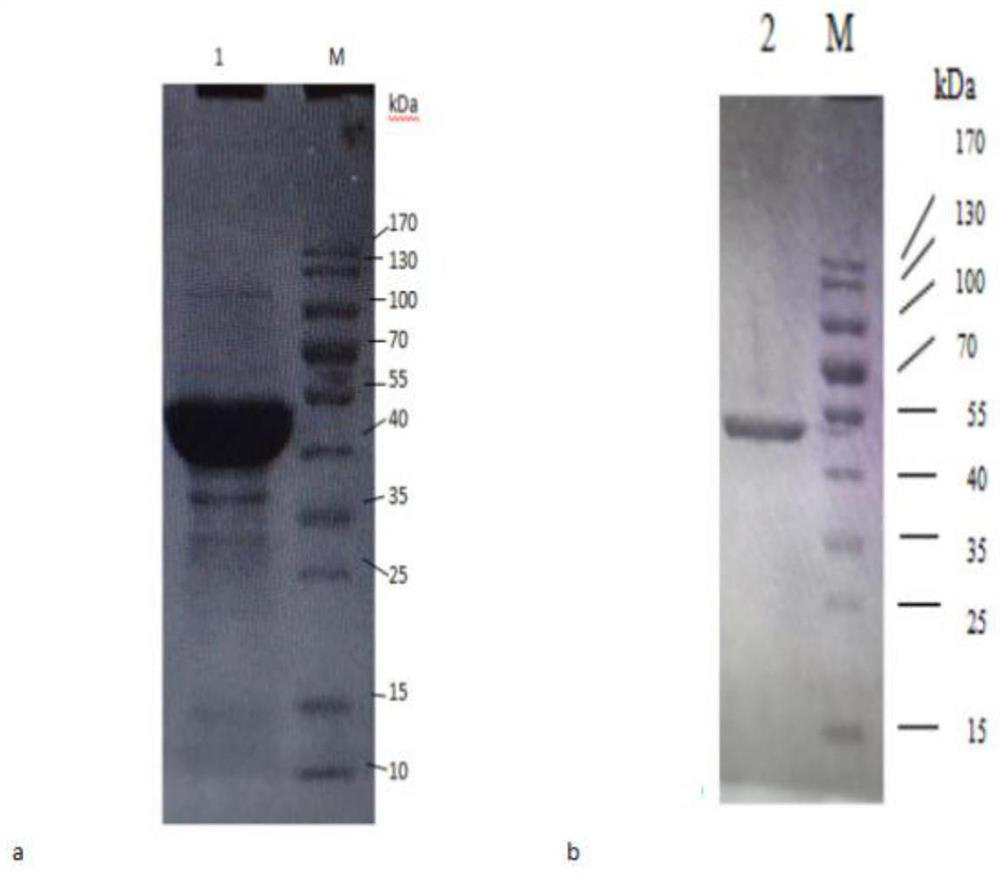
[0063] The purified PCR product and the vector pET-32a(+) were treated with HindIII and BglII double enzyme digestion respectively, and digested at 37° C. for 2...
Embodiment 3
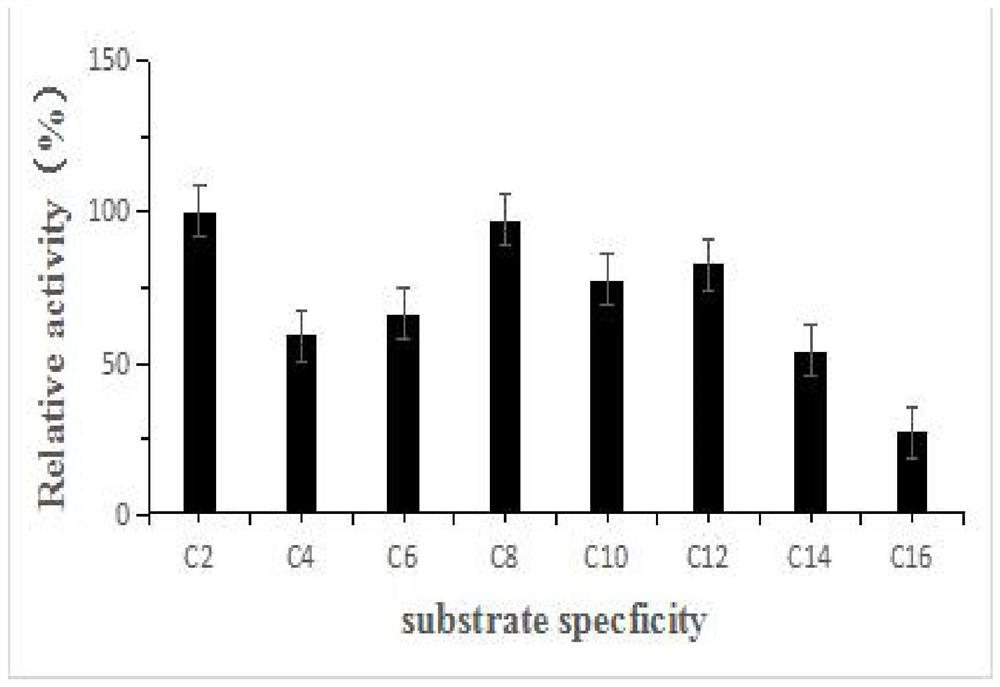
[0077] Embodiment 3: Reaction characteristic of esterase and different substrates
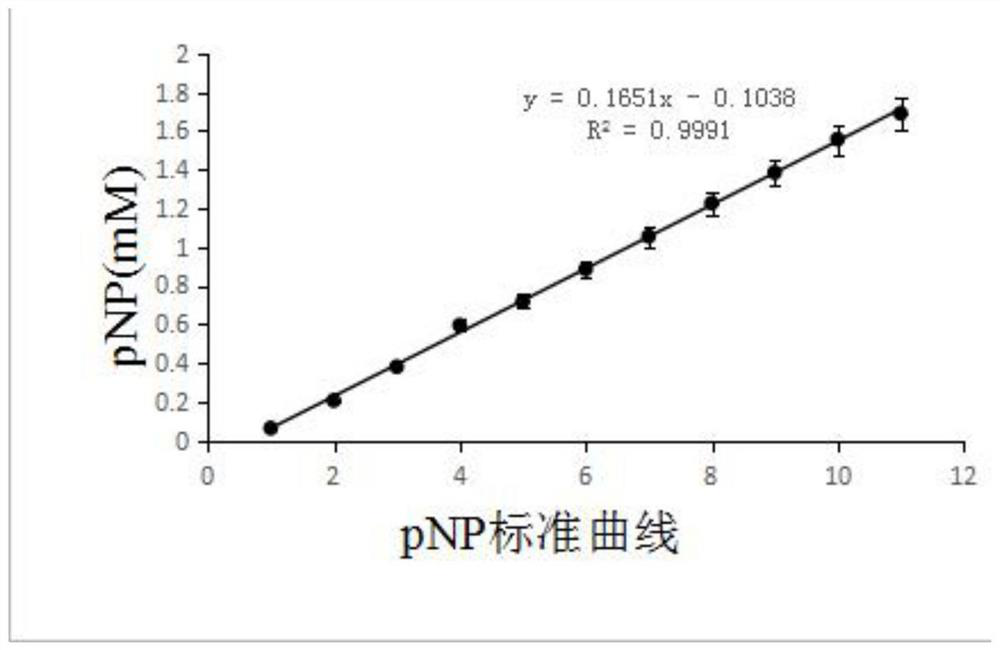
[0078]In order to measure the enzymatic activity of esterase hydrolyzing different carbon chains, according to different enzymatic reaction substrates, 8 types of reaction groups were set up, and each type of reaction group was equipped with three parallel experimental groups and a control experimental group without adding enzyme solution; Each sample in each parallel experimental group is 200 μl enzyme reaction system: 10 μl enzyme solution, final concentration 1mM p-nitrophenol ester (enzymatic reaction substrate), the balance is supplemented with 0.04M B-R tri-acid buffer (pH7), After mixing, react in a water bath at 40°C and pH7 for 20 minutes. In the enzyme reaction system of each parallel experimental group, different substrates with a final concentration of 1 mM were added: p-nitrophenyl acetate (C2), p-nitrophenyl butyrate (C4), p-nitrophenyl caproate (C6), p-nitrophenyl octanoate (C8)...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com