Primer group for detecting PIK3CA gene mutation and application method thereof
A primer set and gene technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problem of low throughput of PIK3CA gene mutation detection, reduce detection cost, improve detection efficiency, The effect of cost saving
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Design and synthesize primer sets
[0065] Step 1.1: According to the sequences of exons 1, 9 and 20 of the PIK3CA gene, design upstream and downstream primers for specifically amplifying gene exons.
[0066] For the design of primers, Primer Quest and Primer Premier 5.0 were used to design primers and analyze dimers and stem-loop mismatches. Primers were designed at both ends containing mutation sites, and the annealing temperatures of the three pairs of primers were basically consistent.
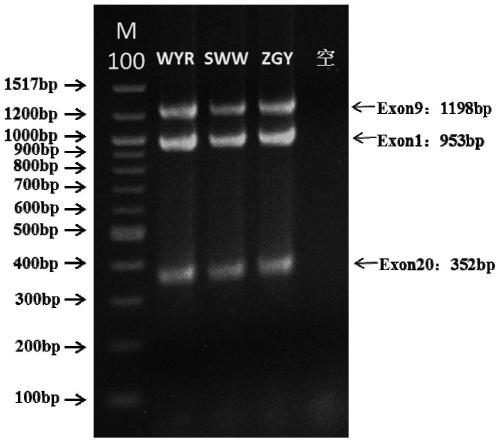
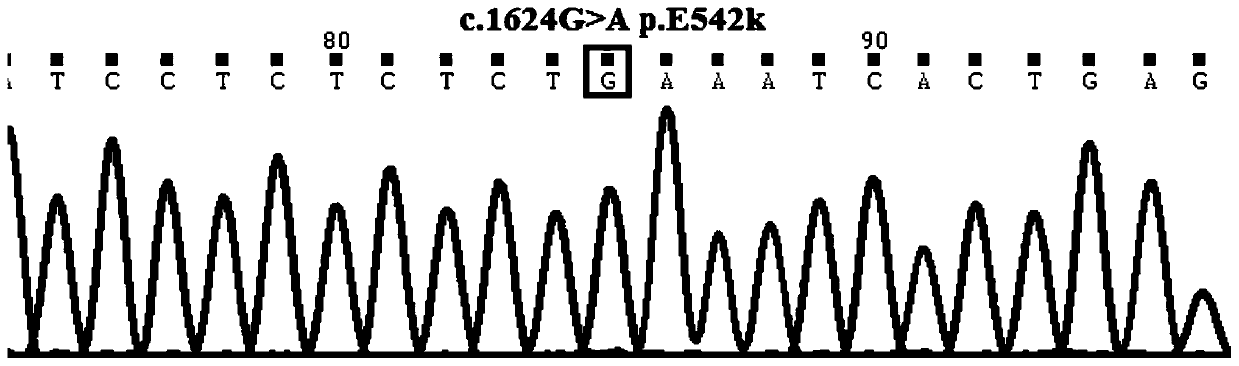
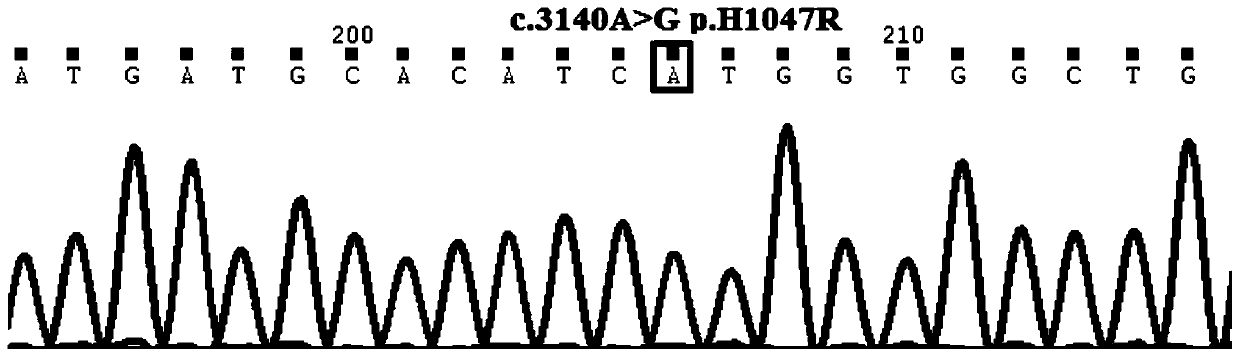
[0067] The primer set provided in this example covers at least one exon of exon 1, exon 9 and exon 20 of the PIK3CA gene. Since small sequence changes will lead to a significant decrease in primer amplification efficiency and poor specificity, multiple PCR primer sets were designed for different sites / exons, and after pre-experimental screening, the length and position of the product fragments were integrated. According to the inclusion situation, the primer set with the best ampli...
Embodiment 2
[0073] Extract DNA from the sample to be tested as an amplification template
[0074] Step 2.1: The sample to be tested can be: serum, plasma sample or collected tumor tissue sample collected from fresh peripheral blood, or oral exfoliated cells collected from a human body with a buccal swab, or fresh peripheral blood collected from a human body.
[0075] Step 2.2: Specifically, use the Tiangen Oral Swab DNA Extraction Kit (DP322), or use the Blood / Cell / Tissue Genomic DNA Extraction Kit (DP304) to extract DNA from the sample, and use the NP80-touch (Germany IMPLEN) measure the concentration and purity of DNA, and preserve the DNA.
Embodiment 3
[0077] Preparation of Multiplex PCR Amplification Reaction System
[0078] Step 3.1: Using the DNA obtained in step 2.2 as an amplification template, and using the primer set synthesized in step 1.2, prepare a multiplex PCR amplification reaction system.
[0079] In this example, the DNA polymerase and buffer in the KOD FX enzyme system (article number: KFX-101) of TOYOBO Co., Ltd. were used as the basic raw materials. concentration, buffer concentration, and enzyme dosage to prepare a multiplex PCR amplification system. The specific composition of this reaction system is shown in Table 3 below.
[0080] It can be understood that the proportional amplification / reduction of the reaction system is within the protection scope of the embodiment of the present invention; the purpose of amplification can also be achieved by replacing other DNA polymerase systems and adjusting the appropriate ratio.
[0081] table 3
[0082]
[0083]
[0084] Primer Mix is a mixture of prim...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com