Construction method and device of benign and malignant tumor identification model
A technology for constructing methods and identifying models, applied in the field of gene detection, can solve the problems of difficulty in taking into account other cancer types, building a benign and malignant tumor discrimination model, and taking into account both broad spectrum and accuracy, and achieves broad-spectrum and specificity. sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0068] Example 1 Construction of tumor benign and malignant differentiation model
[0069] This embodiment provides a method for constructing a model for distinguishing benign from malignant tumors, the flow chart is as follows figure 1 shown, including the following steps:
[0070] (1) Take 20 known benign tumor samples and 65 known malignant tumor samples in Table 1 as the training set; for the above 85 known benign and malignant tumor tissue samples (peripheral blood of tumor samples can also be used, in this In the embodiment, the genomic DNA of the tumor tissue is selected to be amplified by multiplex PCR technology or 5' RACE technology. In this embodiment, the multiplex PCR technology is selected, and the primers used are from the QIAGEN Multiplex PCR Kit, including 32 pairs of forward V Gene primers and 13 pairs of reverse J gene primers, the amplified products were further amplified with Illumina universal primers for the second round to generate a library with a tar...
Embodiment 2
[0097] Example 2 Device for distinguishing benign and malignant tumors
[0098] This embodiment provides a device for distinguishing benign and malignant tumors, the device structure diagram is as follows Figure 4 shown, including:
[0099] The TCR clone identification unit is used to identify the TCR clone type and CDR3 segment of the training set sample;
[0100] CDR3 statistical unit, used to count and calculate the frequency of occurrence of the CDR3 segment;
[0101] Kmer interrupt unit, for recoding the CDR3 segment by Kmer interrupt;
[0102] A data dimensionality reduction unit, used for data dimensionality reduction of the Kmer frequency data after CDR3 recoding;
[0103] The machine learning model training unit is used to associate the benign and malignant information of the known tumor samples in the training set with the Kmer frequency data after data dimensionality reduction, and use the machine learning algorithm to train the model to obtain a benign and mali...
experiment example 1
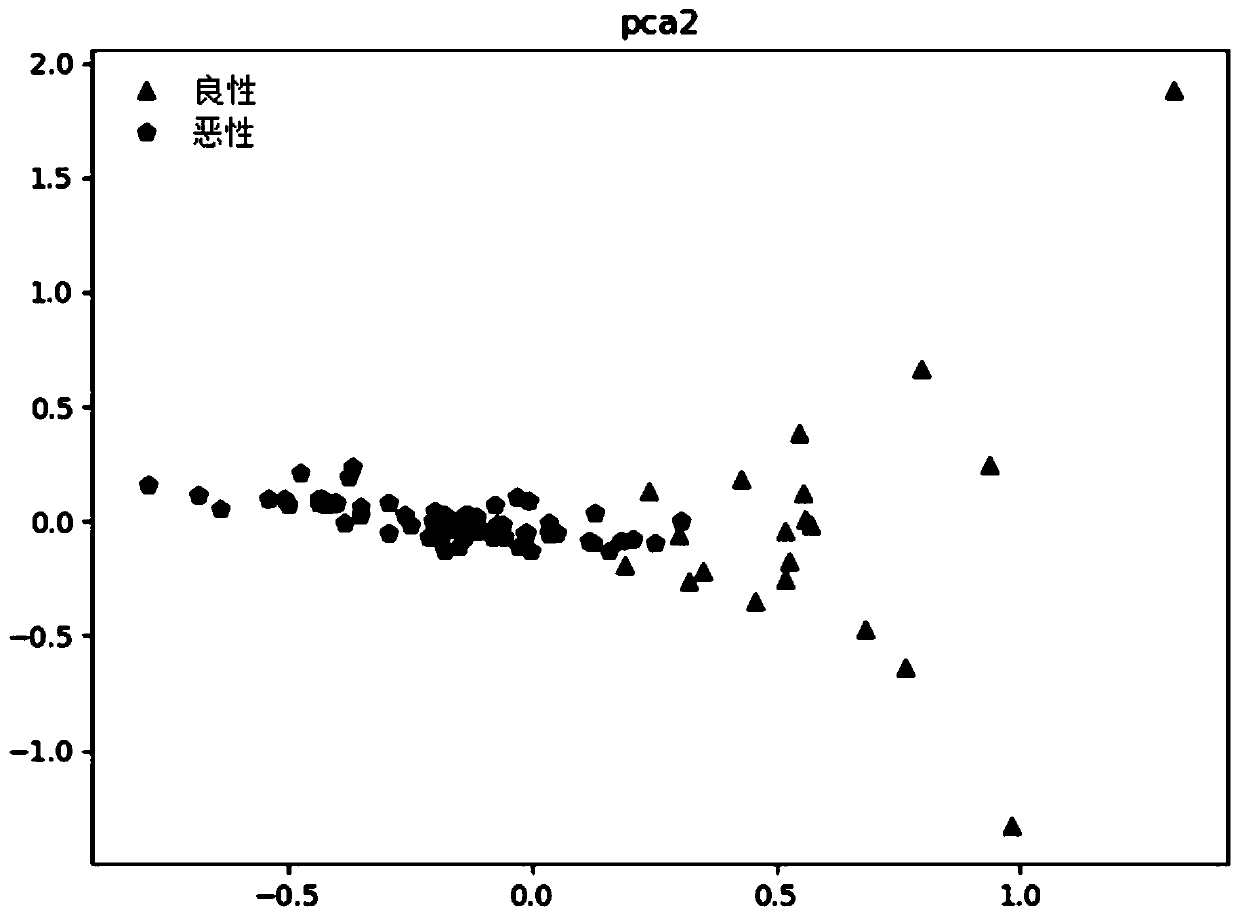
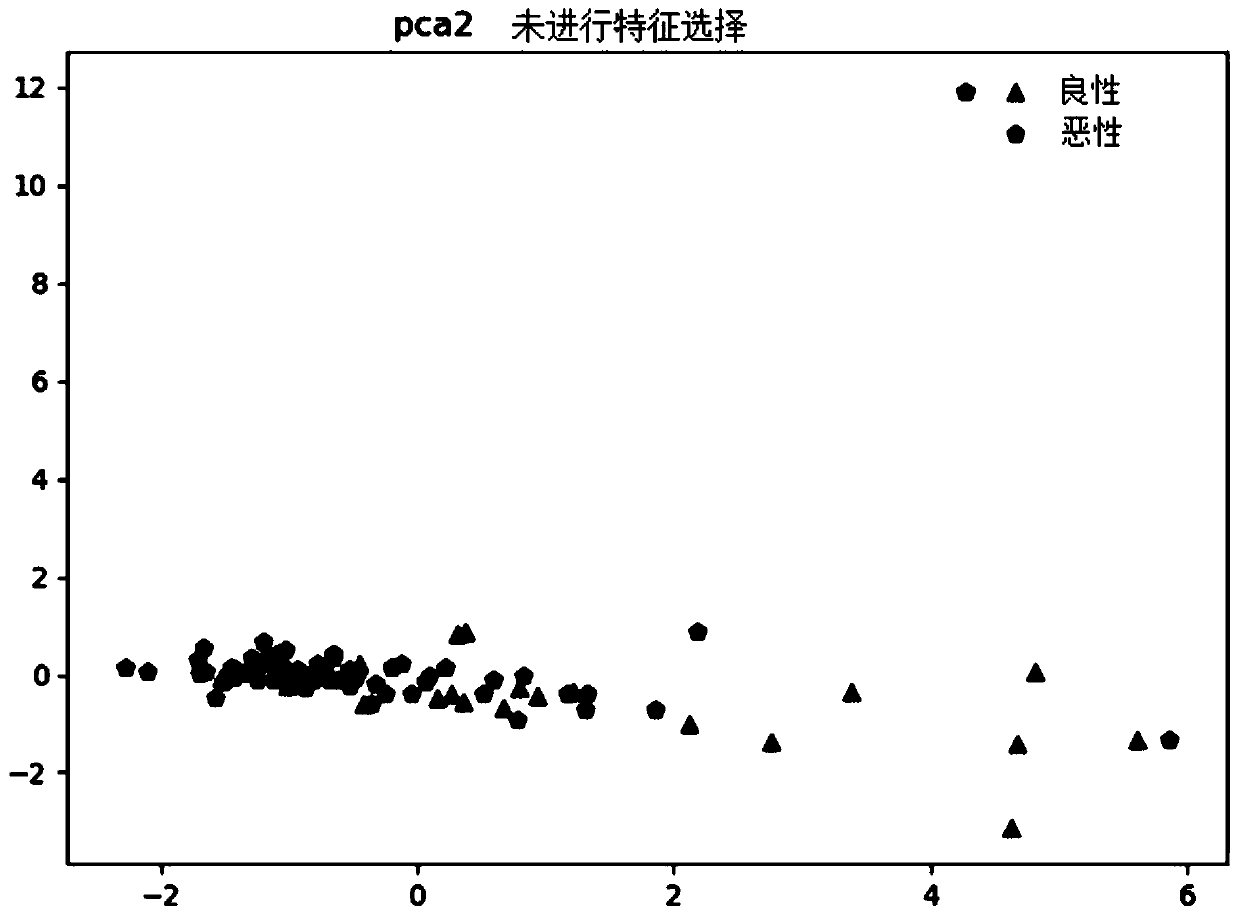
[0119] In this experimental example, the classification model in machine learning is used to perform model training and performance prediction on the Kmer frequency data after dimensionality reduction processing. Specifically, the original data is divided into 5 parts by stratified sampling by means of 50-fold cross-validation. While keeping the ratio of the two types of data for each fold and the original data similar, the model is trained with 4 fold data each time, and the remaining 1 fold data is used to verify the predictive performance of the model. Construction of Example 1 model is verified. The specific implementation is as follows:
[0120] Implement according to Example 1, get 20 known benign tumor samples in Table 1, 65 known malignant tumor samples, as a training set, process the samples according to steps (1)-(6) in Example 1, and The Kmer frequency data obtained after dimensionality reduction adopts the method of 5-fold cross-validation to divide the original d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com