Molecular docking method based on polarizable bond model
A molecular docking and modeling technology, which is used in the analysis of two-dimensional or three-dimensional molecular structure, molecular design, computational theoretical chemistry, etc. It can solve the problem of not considering the polarization effect, and achieve the effect of improving the accuracy of molecular docking.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
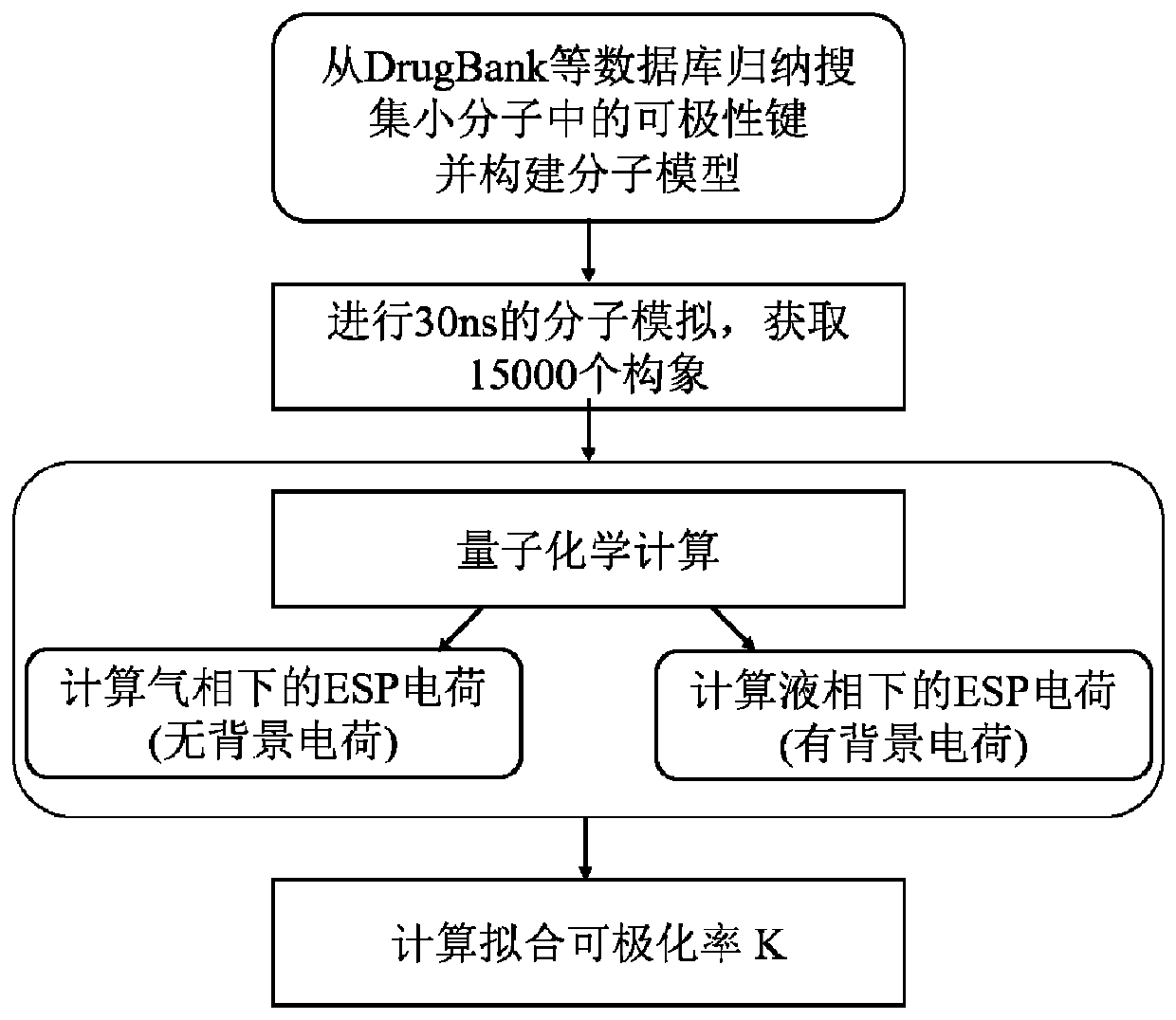
[0052] See attached Figure 1 ~ Figure 2 , define the type of polarizable bond in the compound and calculate the polarization parameter of the polarizable bond according to the following steps:
[0053] 1) Collect the types of polarizable bonds from Drugbank, ChemBL and other databases, then construct a simplified molecular model, and obtain the polarizability and equilibrium bond length of small molecule polarizable bonds by combining molecular simulation and quantum chemistry;
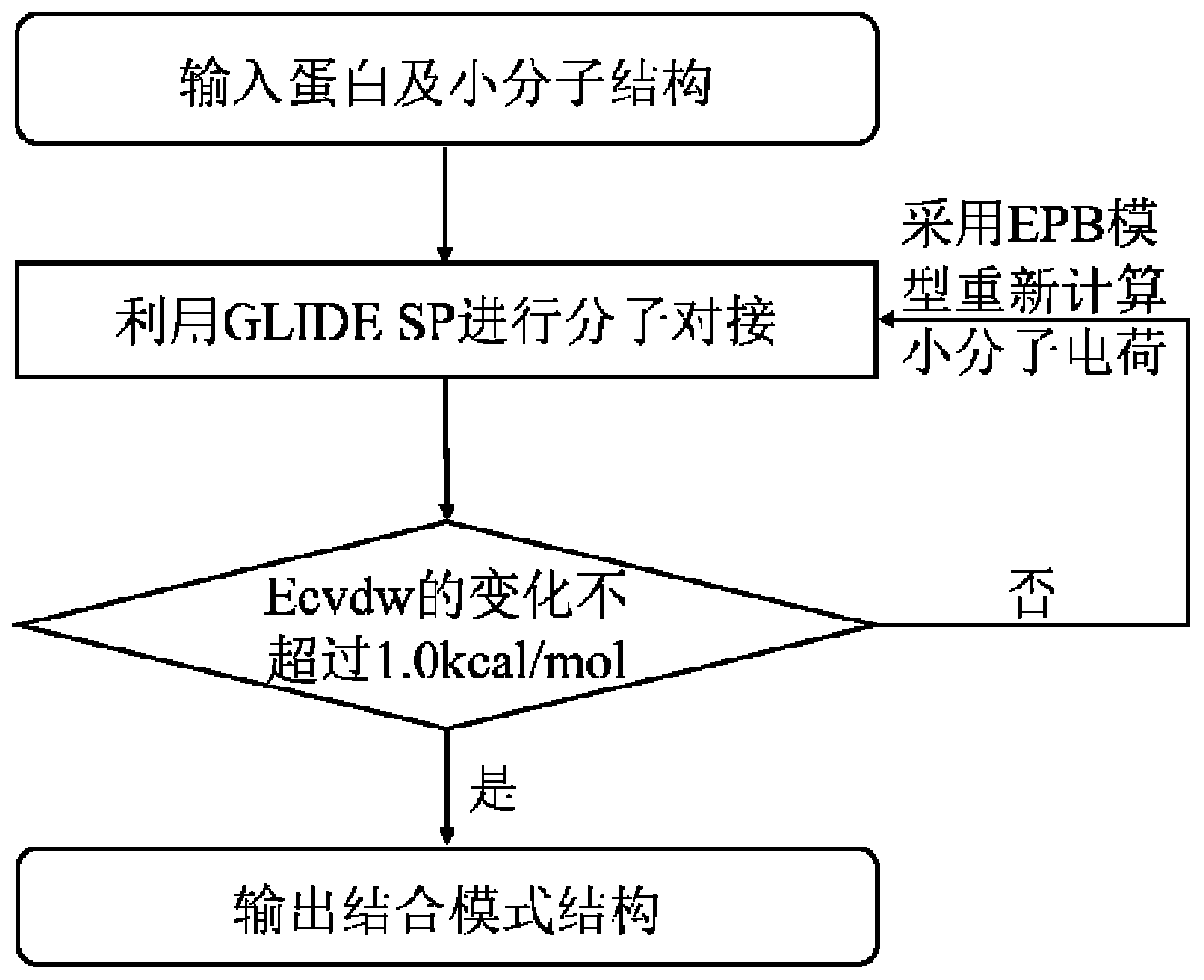
[0054] 2) Download the complex and define the binding pocket;
[0055] 3) Molecular docking was performed using the Schrödinger molecular docking software Glide;
[0056] 4) Based on the small molecule EPB polarized charge model, each molecule after docking recalculates the charge in its protein environment, and then re-docks the molecule, and the updated charge is forced to be used during the docking process;
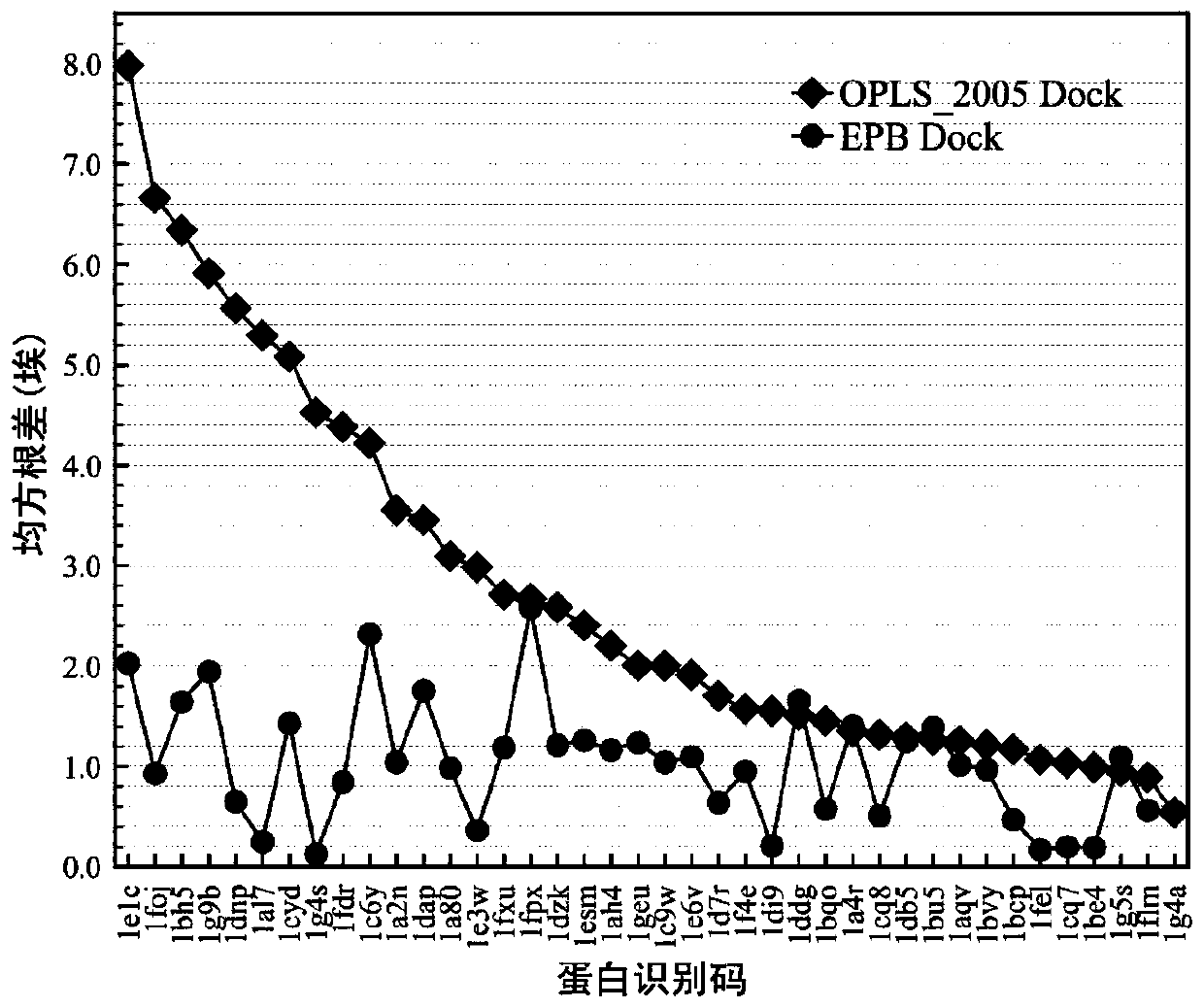
[0057] 5) Repeat step 4 to repeatedly carry out the molecular docking of the EPB model ...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap