Method and system for constructing models for predicting protein-RNA interaction binding sites
A binding site and protein technology, applied in the field of RNA-protein interaction prediction, can solve the problems of model prediction accuracy, increased calculation time, overfitting, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
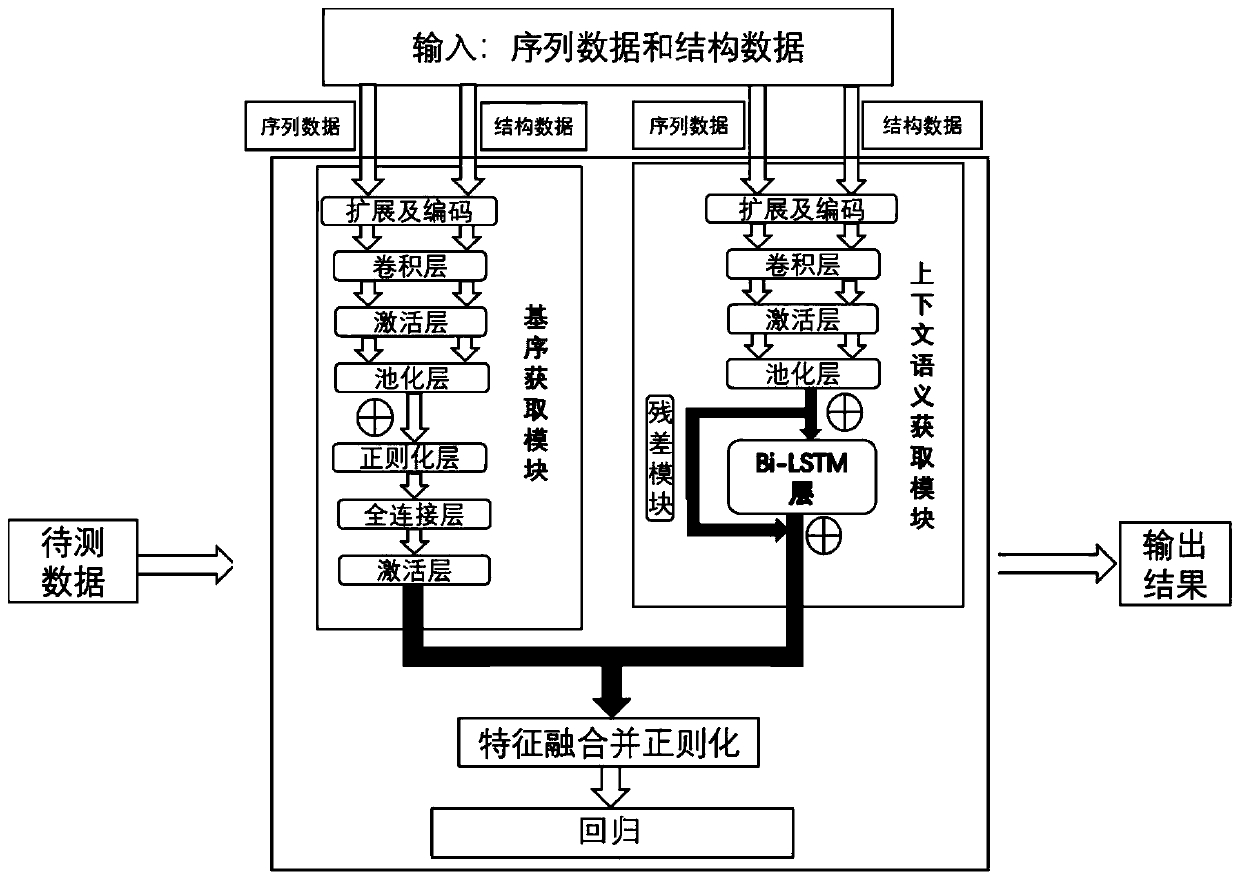
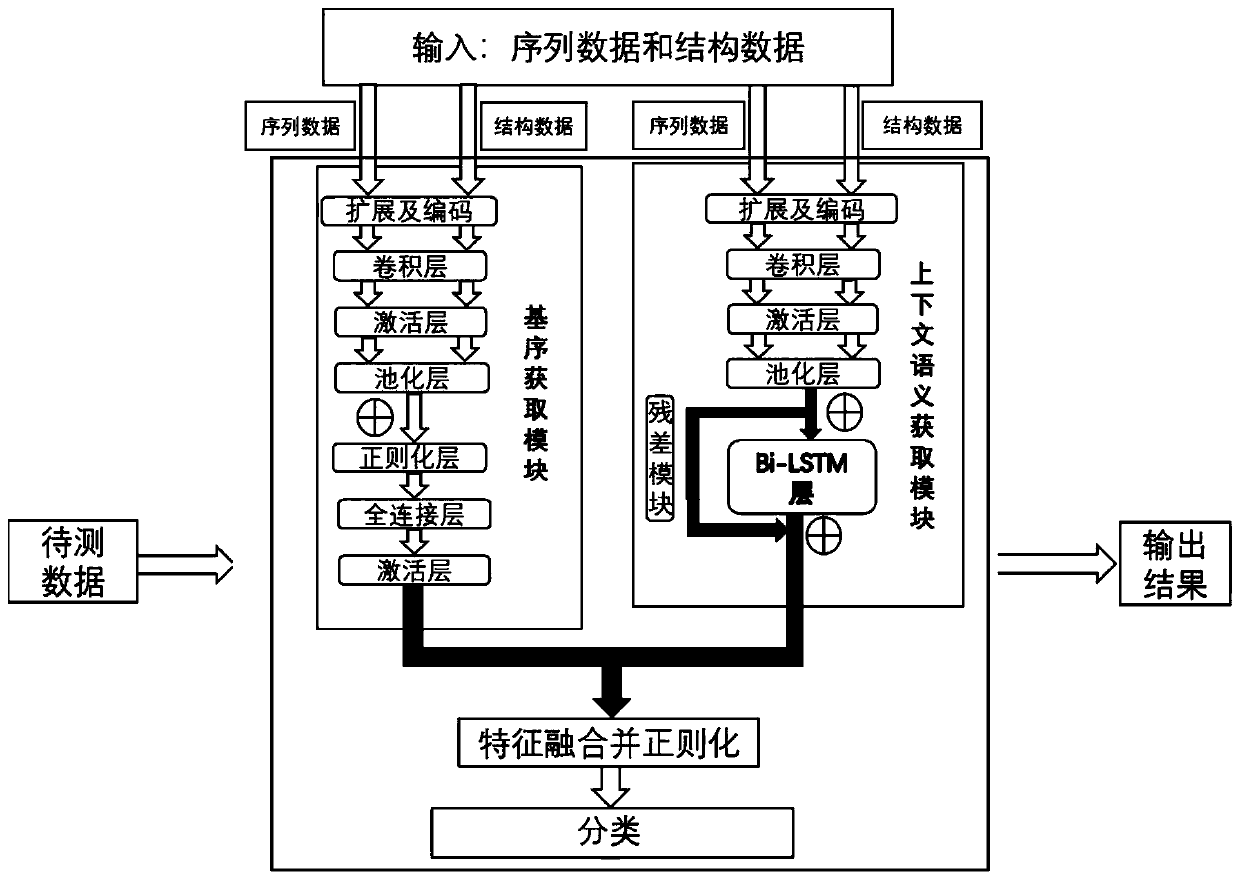
[0050] According to one aspect of the present invention (RBPnet), the original data used for training the model not only includes the sequence data of the RNA bound to the protein, but also introduces the structural omics measurement data generated by the RNA structural omics measurement experimental technology, such as DMS-seq structural omics determination data were used as input data for the model. Compared with the existing technologies (such as the models such as Deepnet-rbp mentioned above), the RNA structural omics data based on DMS-seq technology can provide the secondary structure information of RNA in the real cell state in vivo, so the present invention uses it In the study of RNA-protein interaction, it is used to solve the problem that the prediction of RNA structure by software is not accurate and cannot reflect the real RNA structure information in vivo.
[0051] According to one aspect of the present invention (RBPnet), two modules are designed in the processin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com