RNA-RP RT-PCR method capable of fast detecting melon viruses
A technology for melons and viruses, applied in the directions of DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., to achieve the effect of saving cost and time, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
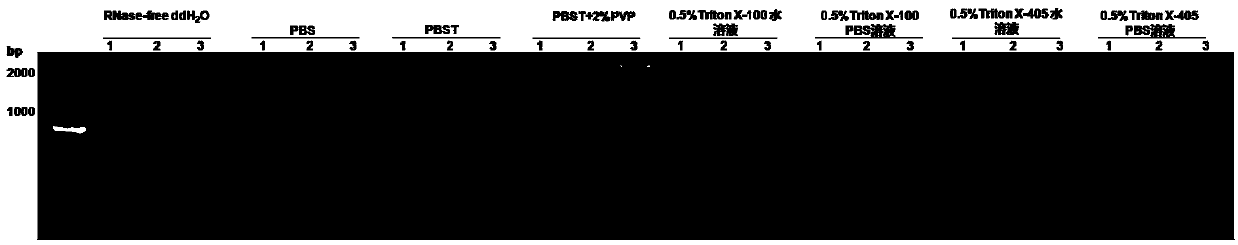
[0050] The screening of solution used in the quick release of embodiment 1 plant total RNA
[0051] Weighed 8 copies of 0.05 g of CGMMV melon diseased leaves, put them in 2 mL centrifuge tubes, and crushed them with liquid nitrogen at low temperature using a tissue disruptor, and added 1 mL of RNase-free ddH2O, 1×PBS, PBST, PBST+2%PVP, 0.5% Triton X-100 aqueous solution, 0.5% Triton X-100 PBS solution, 0.5% Triton X-405 aqueous solution and 0.5% Triton X-405 PBS solution, shake and mix vigorously, 8000 rpm Centrifuge for 5 min, absorb the supernatant for later use;
[0052] Use the CGMMV-specific primer sequence CG-3467F / CG-5294R to carry out virus detection on the prepared supernatant, in which 2×1 Step Buffer (Dye PLus) 10 μL, PrimeScript 1 Step Enzyme Mix 0.8 μL, CG-3467F (10 μM) 1 μL, CG-5294R (10 μM) 1 μL, supernatant 0.5 μL, rehydration 6.7 μL, total PCR system 20 μL; 50°C for 30 min, 94°C for 2 min, 94°C for 30 sec, 57°C for 30 sec, 72°C 2 min, run 35 cycles, total ex...
experiment example 2
[0055] Experimental example 2 RNA-RP RT-PCR detection of CGMMV in tobacco and cucumber
[0056] Weigh two copies of 0.05 g of CGMMV tobacco leaves and cucumber diseased leaves, put them in 2 mL centrifuge tubes, crush them with liquid nitrogen at low temperature with a tissue breaker, and add 1 mL of PBST+2% to each tube sample PVP and 0.5% Triton X-405PBS solution, shake vigorously, mix well, centrifuge at 8000 rpm for 5 min, absorb the supernatant, and use the CGMMV-specific primer sequence CG-3467F / CG-5294R to carry out virus detection on the prepared supernatant, wherein 2×1 Step Buffer (Dye PLus) 10 μL, PrimeScript 1 Step Enzyme Mix 0.8 μL, CG-3467F (10 μM) 1 μL, CG-5294R (10 μM) 1 μL, supernatant 0.5 μL, water 6.7 μL, total A total of 20 μL of PCR system; 35 cycles of 30 min at 50°C, 2 min at 94°C, 30 sec at 94°C, 30 sec at 57°C, 2 min at 72°C, and a total extension of 2 min at 72°C;
[0057] 4 μL of PCR products were taken for electrophoresis detection, and the results...
experiment example 3
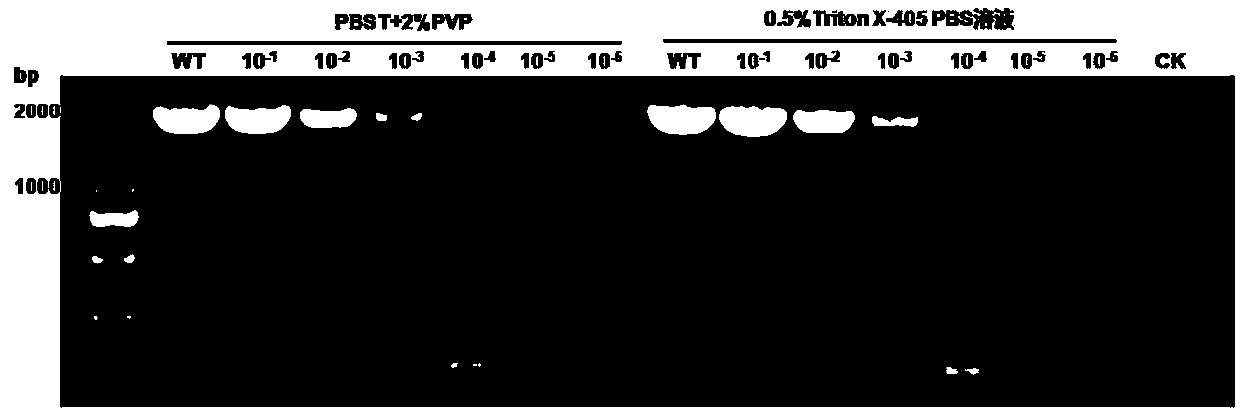
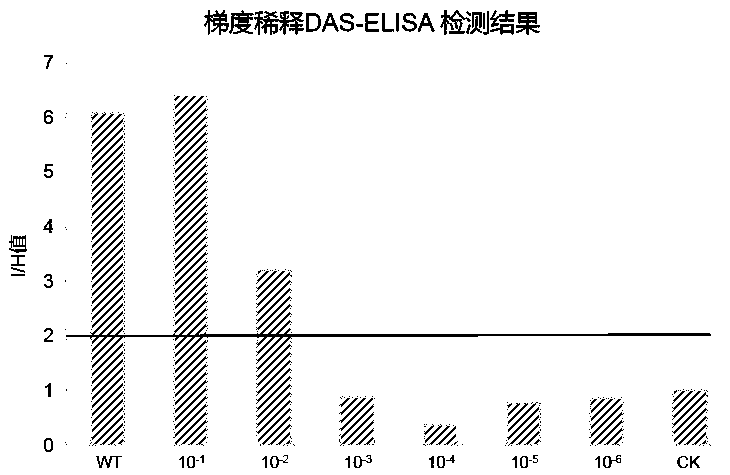
[0058] Experimental example 3 RNA-RP RT-PCR detection of CMV in tobacco and melon
[0059] Weigh 0.05 g of CMV benthamiana benthamiana and melon diseased leaves respectively, put them in 2mL centrifuge tubes, crush them with liquid nitrogen at low temperature with a tissue breaker, and add 1 mL of PBST+2%PVP to each tube sample and 0.5% Triton X-405PBS solution, shake vigorously, mix well, centrifuge at 7000 rpm for 8min, absorb the supernatant, and use specific primers CMV-R1-1F / CMV-R1-1627R combined with a one-step RT-PCR method to detect CMV, Concrete step is the same as embodiment 1, is not repeating,
[0060] 3 μL of PCR products were taken for electrophoresis detection. The results are shown in 5, indicating that both PBST+2%PVP and 0.5% Triton X-405PBS solutions can effectively release the total RNA in Nicotiana benthamiana and melon leaves for the detection of CMV.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com