A method for improving gene replacement efficiency in plants
A gene replacement and plant technology, applied in botany equipment and methods, biochemical equipment and methods, plant products, etc., to achieve the effect of less by-products, high efficiency and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0097] Example 1. Construction of a carrier for gene replacement without DNA double-strand breaks and its application in rice gene replacement
[0098] 1. Construction of recombinant expression vector and description of replacement principle
[0099] 1. Construction of recombinant expression vector
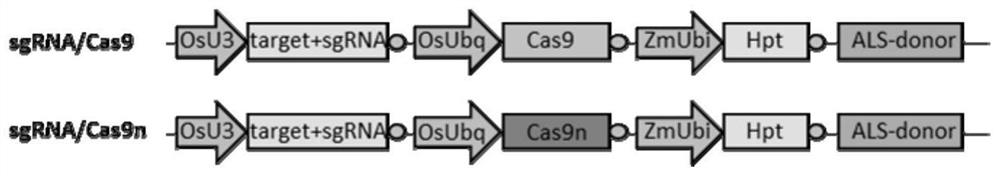
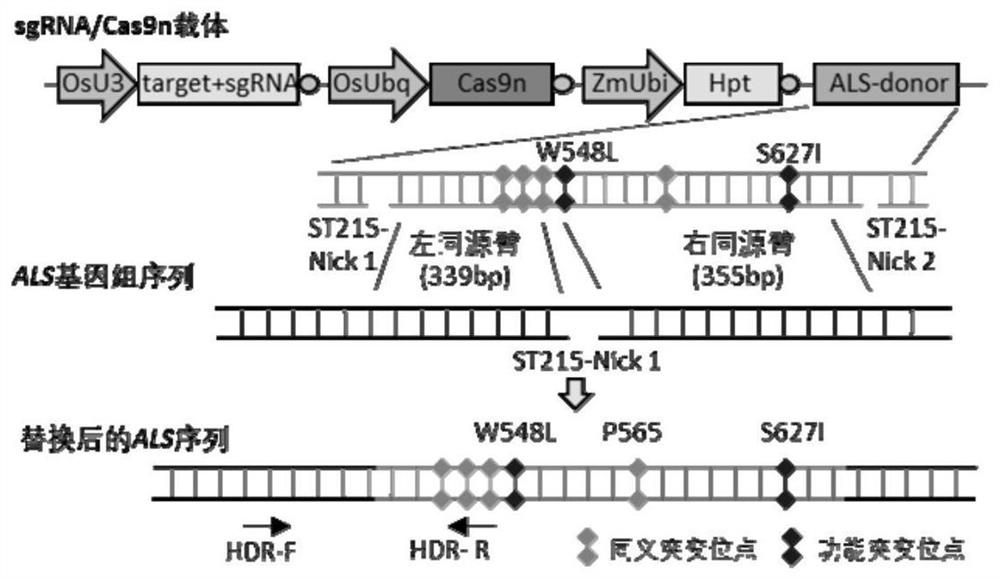
[0100] Artificially synthesize the following recombinant expression vectors, each of which is a circular plasmid: sgRNA / Cas9 recombinant expression vector, sgRNA / Cas9n recombinant expression vector. The structural schematic diagrams of sgRNA / Cas9 recombinant expression vector and sgRNA / Cas9n recombinant expression vector are as follows: figure 1 shown. The specific structure description is as follows:
[0101] The sequence of the sgRNA / Cas9n recombinant expression vector is sequence 1 in the sequence list. The 131-467th position of sequence 1 is the OsU3 promoter sequence, the 474-550th position is the tRNA sequence, the 551-570th position is the ST215 target sequence, the 571...
Embodiment 2
[0145] Example 2, Optimization of sgRNA / Cas9n recombinant expression vector and its application in rice gene replacement
[0146] 1. Construction of recombinant expression vector and description of replacement principle
[0147] 1. Construction of recombinant expression vector
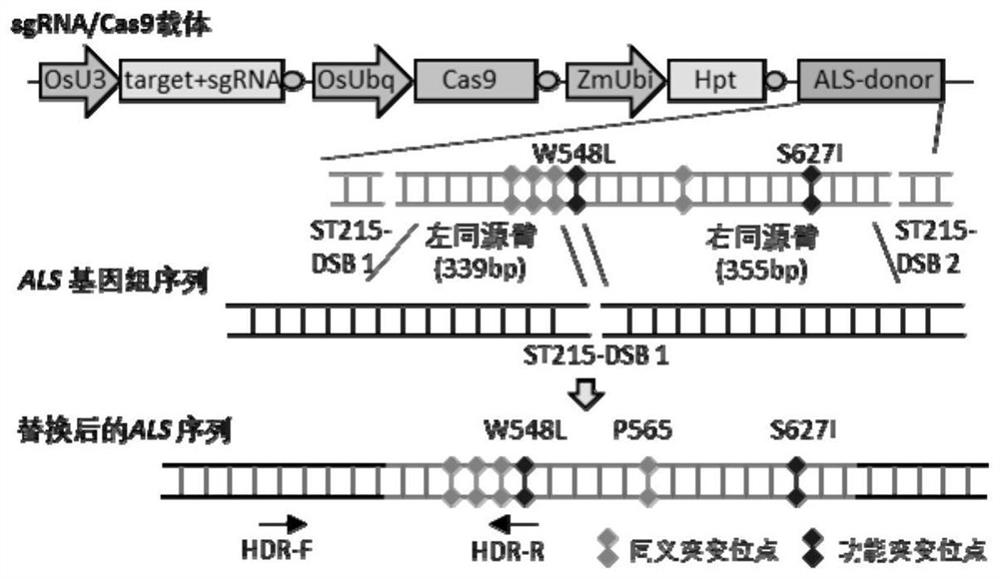
[0148] Artificially synthesize the following recombinant expression vectors, each of which is a circular plasmid: sgRNA / Cas9n recombinant expression vector, esgRNA / Cas9n-P2A-Hpt recombinant expression vector. Schematic diagram of the structure of sgRNA / Cas9n recombinant expression vector and esgRNA / Cas9n-P2A-Hpt recombinant expression vector Figure 7 shown. The specific structure description is as follows:
[0149] The sequence of the sgRNA / Cas9n recombinant expression vector is sequence 1 in the sequence list.
[0150] The sequence of the esgRNA / Cas9n-P2A-Hpt recombinant expression vector is to replace the 571-646 in the sequence 1 with the esgRNA backbone sequence shown in the sequence 7, and re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com