Glypican-3 peptide reagents and methods
A phosphatidylinositol and proteoglycan technology, applied in the field of glypican-3 peptide reagents and methods, can solve the problems of shortened survival time and low effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
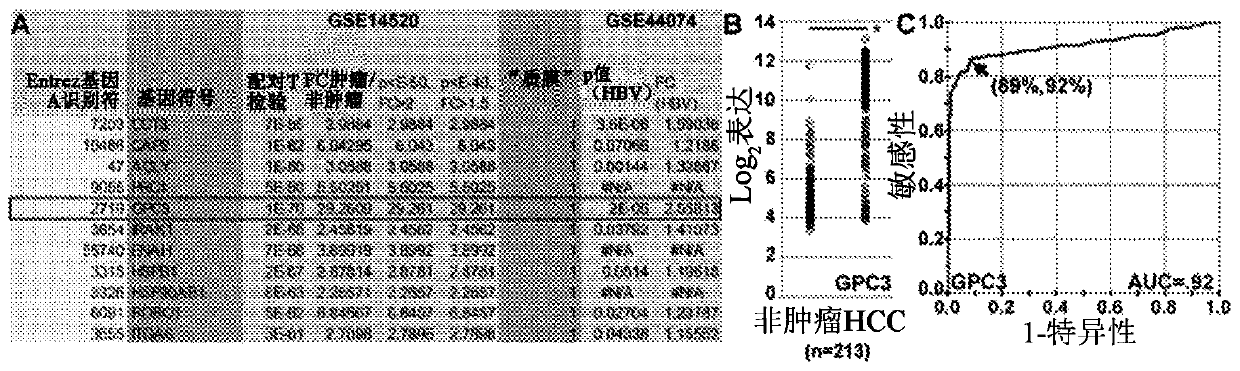
[0097] GPC3 expression
[0098] We use datasets GSE14520 [Roessler et al., Cancer Res., 70:10202-10212 (2010)] and GSE44074 [Ueda et al., Genomics, 101:238-248 (2013 )] identified GPC3 as a promising target for detection and treatment of HCC. Expression levels of GPC3 showed large differences in P-values and mean fold changes compared to non-tumors, Picture 1-1 a. difference by log 2 Displays the distribution of gene expression levels to reflect, as Picture 1-1The dataset GSE14520 in B is shown. Figure 1-2 B display with Picture 1-1 The same dataset GSE14520 is shown in B, and an additional dataset GSE44074 is shown (ie both datasets are shown). The ROC curve for this data showed a sensitivity of 89% and a specificity of 92%, with an area under the curve (AUC) of 0.92, Picture 1-1 c. These results suggest that GPC3 is a promising target for HCC. Figure 1-2 A is shown under additional analysis with Picture 1-1 The same data as shown in A. Figure 1-2 C is si...
example 2
[0101] Peptides specific for GPC3
[0102] A panel of candidate peptides specific for GPC3 was identified using phage display technology. Peptide selection was performed using an M13 phage library expressing about 10 of each individual sequence. 9 unique clones [Zhou et al., Clin Transl Gastroenterol, 6:e101 (2015); Joshi et al., Bioconjug Chem, 27:481-494 (2016); Rabinsky et al., Cell Mol Gastroenterol Hepatol, 2:222-237 (2016)]. Biopanning of the library was performed against purified recombinant GPC3 core protein immobilized in 6-well plates. Four rounds of biopanning were performed using decreasing amounts (100, 80, 60 and 40 μg) of GPC3 core protein in successive rounds to increase binding specificity. Bound phage were eluted, amplified, precipitated, and titrated using standard protocols, and enriched clones from the candidate pool were sequenced to identify lead candidate sequences: ALLANHEELF (SEQ ID NO:2), GLHTSATNLYLH (SEQ ID NO:3 ), SGVYKVAYDWQH (SEQ ID NO: 4) a...
example 3
[0107] siRNA knockdown of GPC3
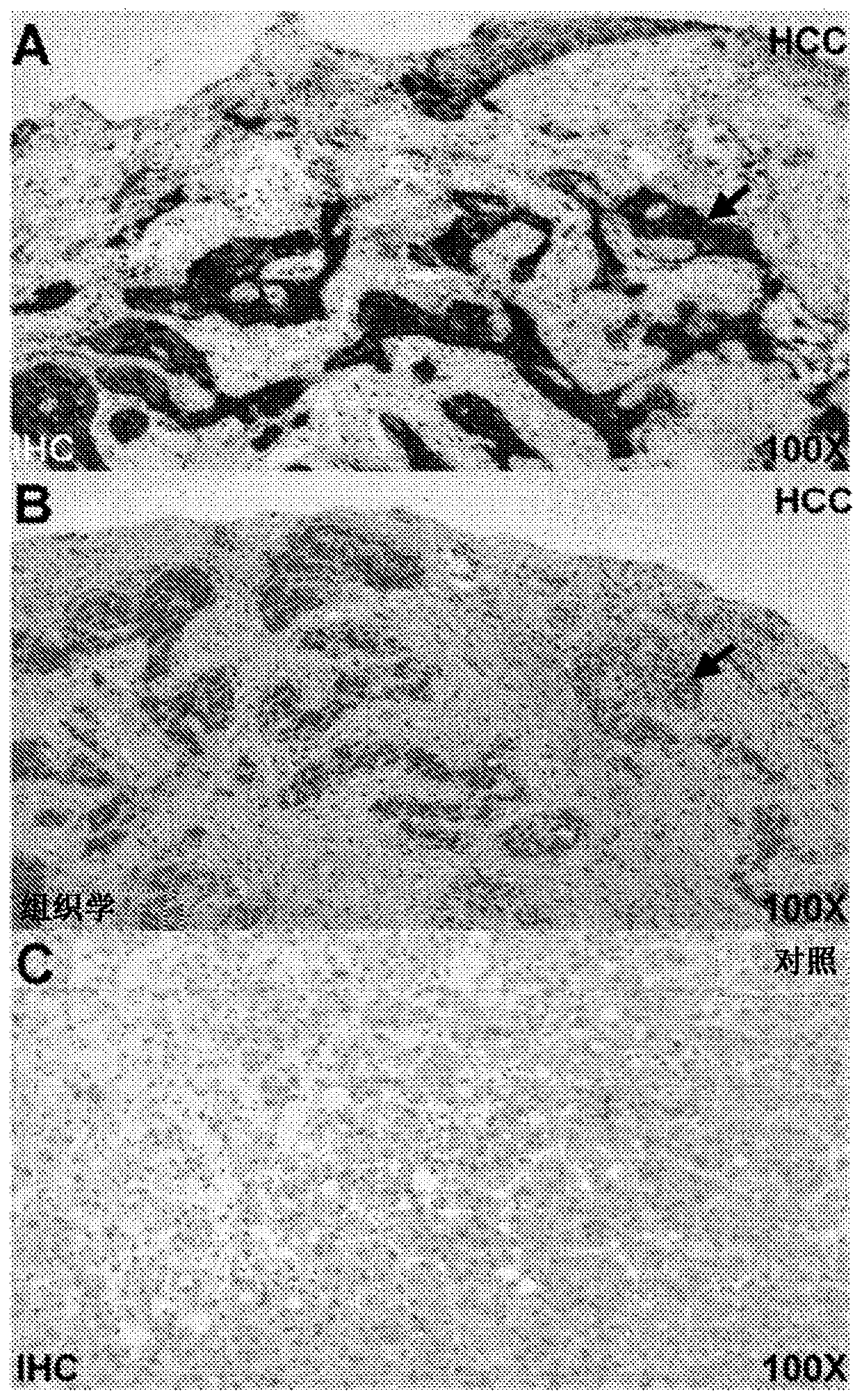
[0108] The specificity of lead candidate peptide reagent binding was verified by siRNA knockdown of GPC3 in human HCC cells. Cells were transfected with siRNA to knockdown the cell surface expression of GPC3. In confocal microscopy, Figure 6-1 A shows ALL*-Cy5.5 peptide reagent, and Figure 6-1 B shows strong binding of AF488-labeled anti-GPC3 antibody to the cell surface of Hep3B cells (arrows). These cells were transfected with non-targeting siRNA (siCL) to serve as controls. Figure 6-1 C, F show minimal binding of the scrambled (control) peptide QLE*-Cy5.5 to the same cells. Figure 6-1 D and E show that for Hep3B cells transfected with siRNA (siGPC3) to knock down the expression of GPC3, the fluorescence intensity of peptide reagent and anti-GPC3 antibody decreased about 4 times, respectively. Figure 6-1 G shows measurements of fluorescence intensity for images collected in triplicate. Figure 6-1 H shows western blot of GPC3 expre...
PUM
| Property | Measurement | Unit |
|---|---|---|
| affinity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com