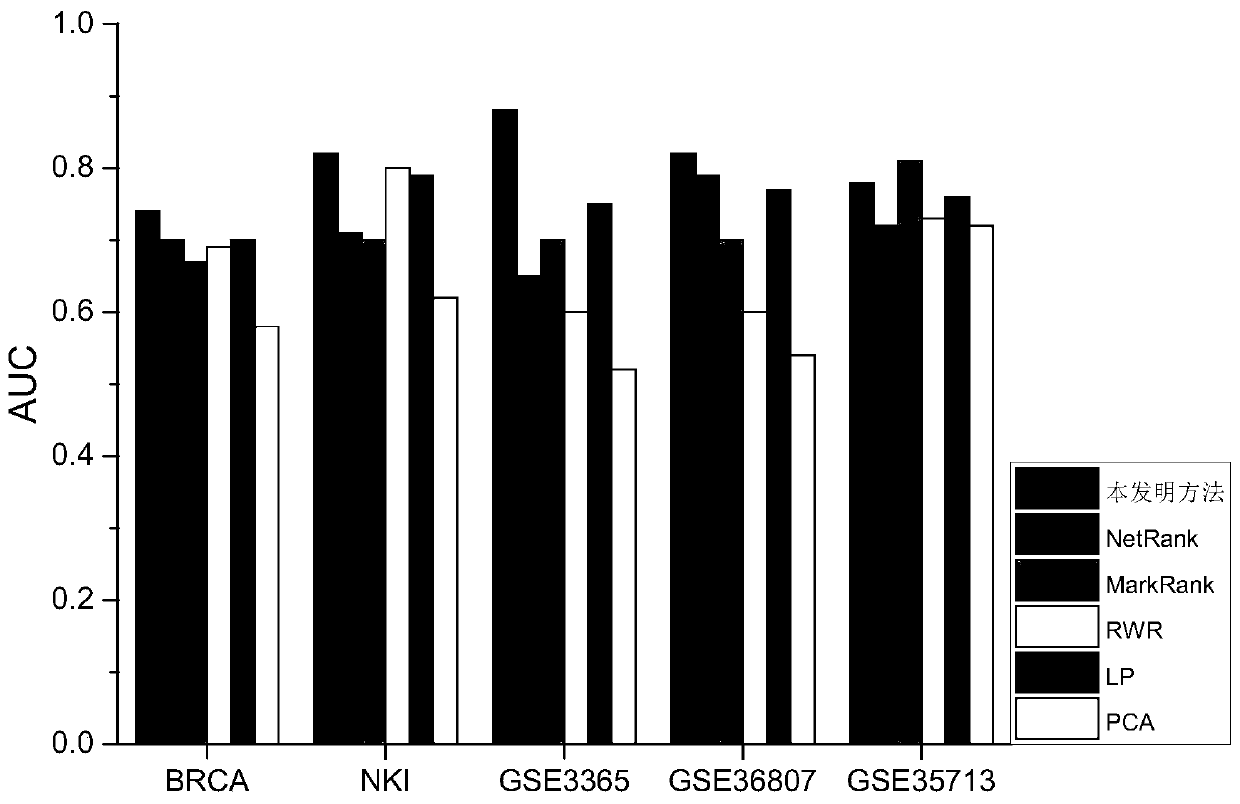
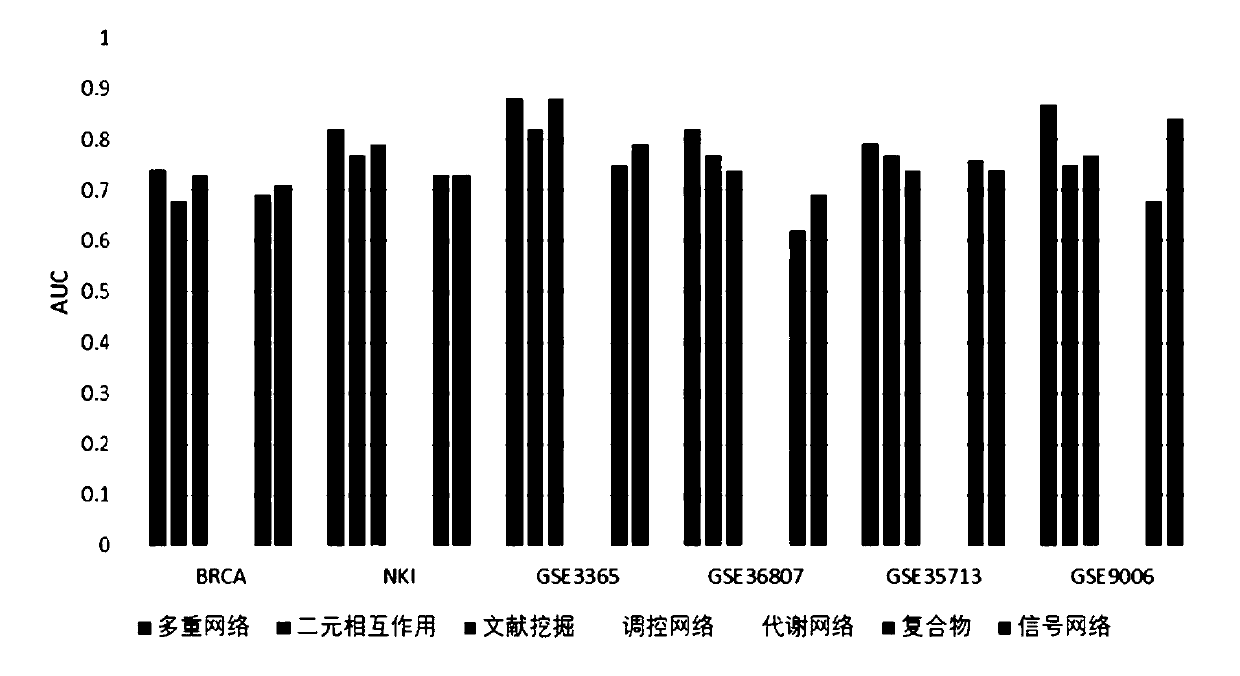
Biomarker identification method based on multiple networks
A technology of biomarkers and identification methods, applied in the fields of biological systems, bioinformatics, neural learning methods, etc., can solve problems such as the inability to reflect the characteristics and topology structure well, and the mutual interference of data.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
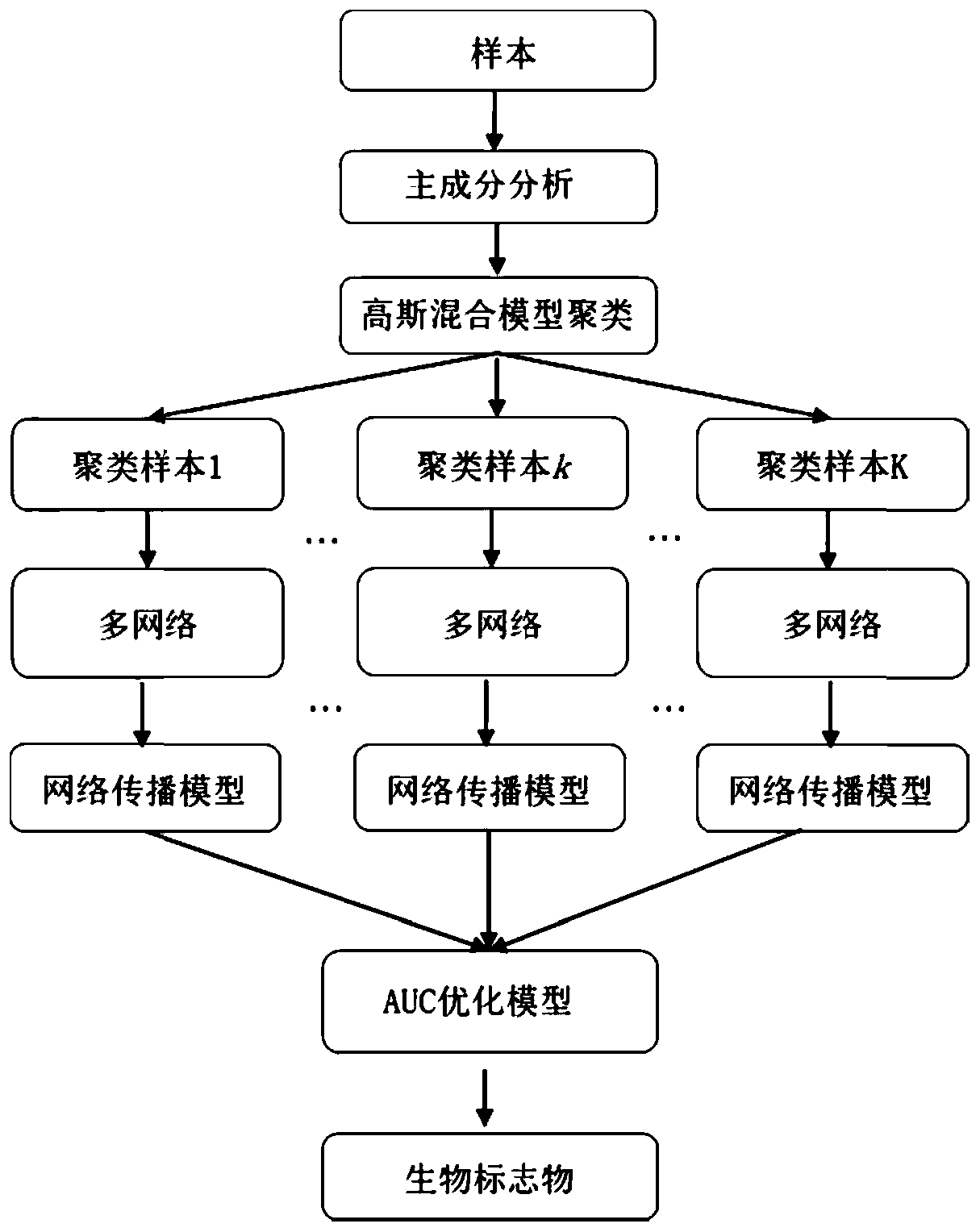
[0027] 1. Preprocessing of gene expression data
[0028] Read in a gene expression data file and normalize the gene expression data by Z-score:
[0029]
[0030] x represents the original gene expression value of each sample; μ represents the mean of all gene original expression data of each sample; σ is the standard deviation of all gene original expression data of each sample.
[0031] 2. Principal component analysis of gene expression data
[0032] Based on the standardized gene expression data, the specific process of obtaining the first two principal components of the gene expression matrix through principal component analysis is as follows:
[0033] 1) Find the covariance matrix of the features in the standardized gene expression data;
[0034] 2) Find the eigenvalues and corresponding eigenvectors of the covariance matrix;
[0035] 3) Sort the eigenvalues in descending order, select the two largest ones, and then use the two corresponding eigenvectors as colum...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com