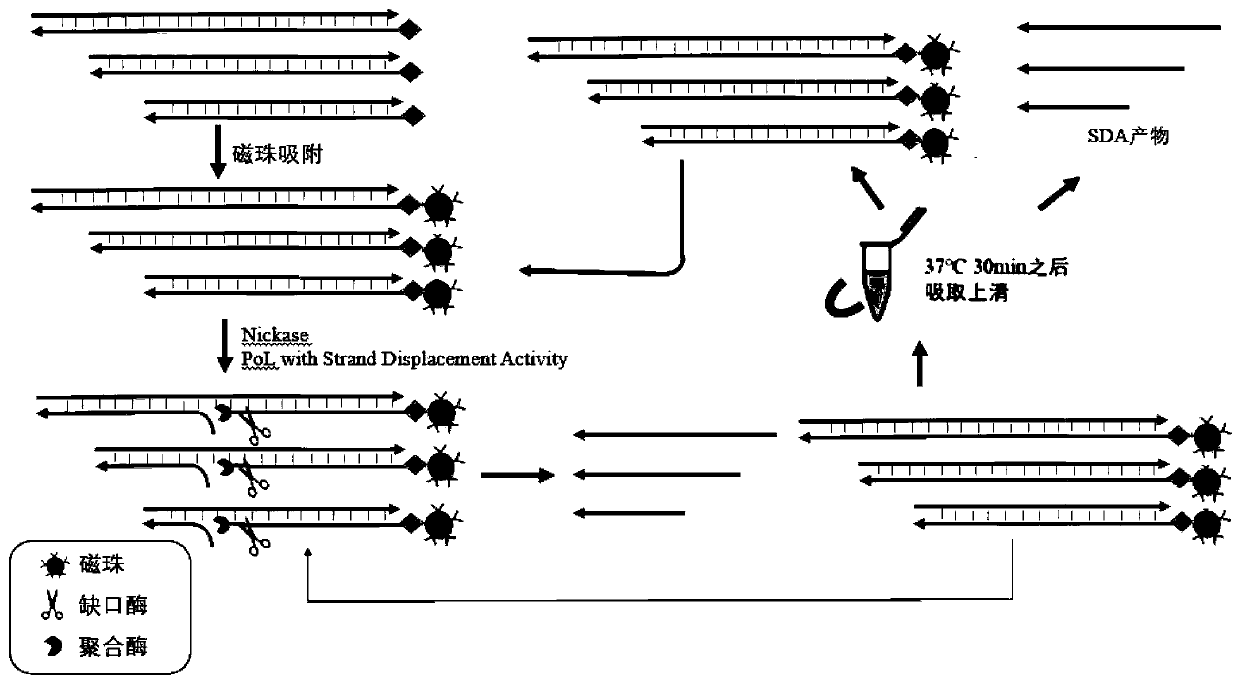
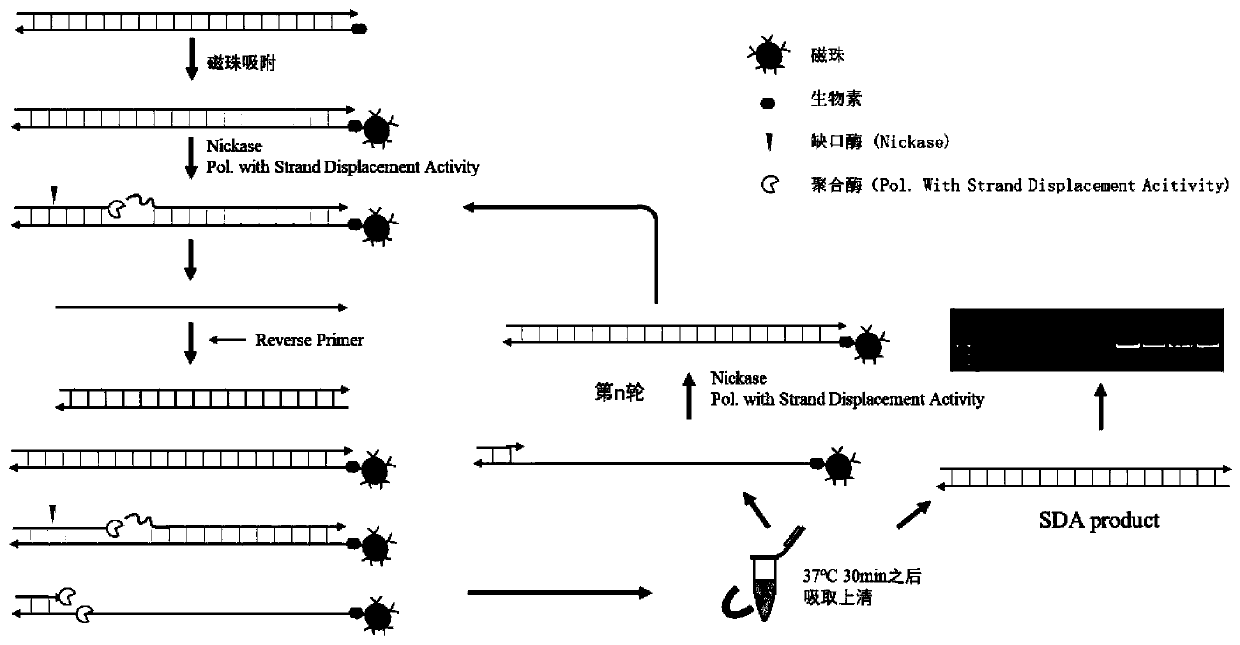
Isothermal oligonucleotide library amplification method applied to DNA data storage
An oligonucleotide and constant temperature amplification technology, applied in the field of constant temperature amplification of oligonucleotide libraries, can solve the problems of accelerated DNA decay, data loss, and increased costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
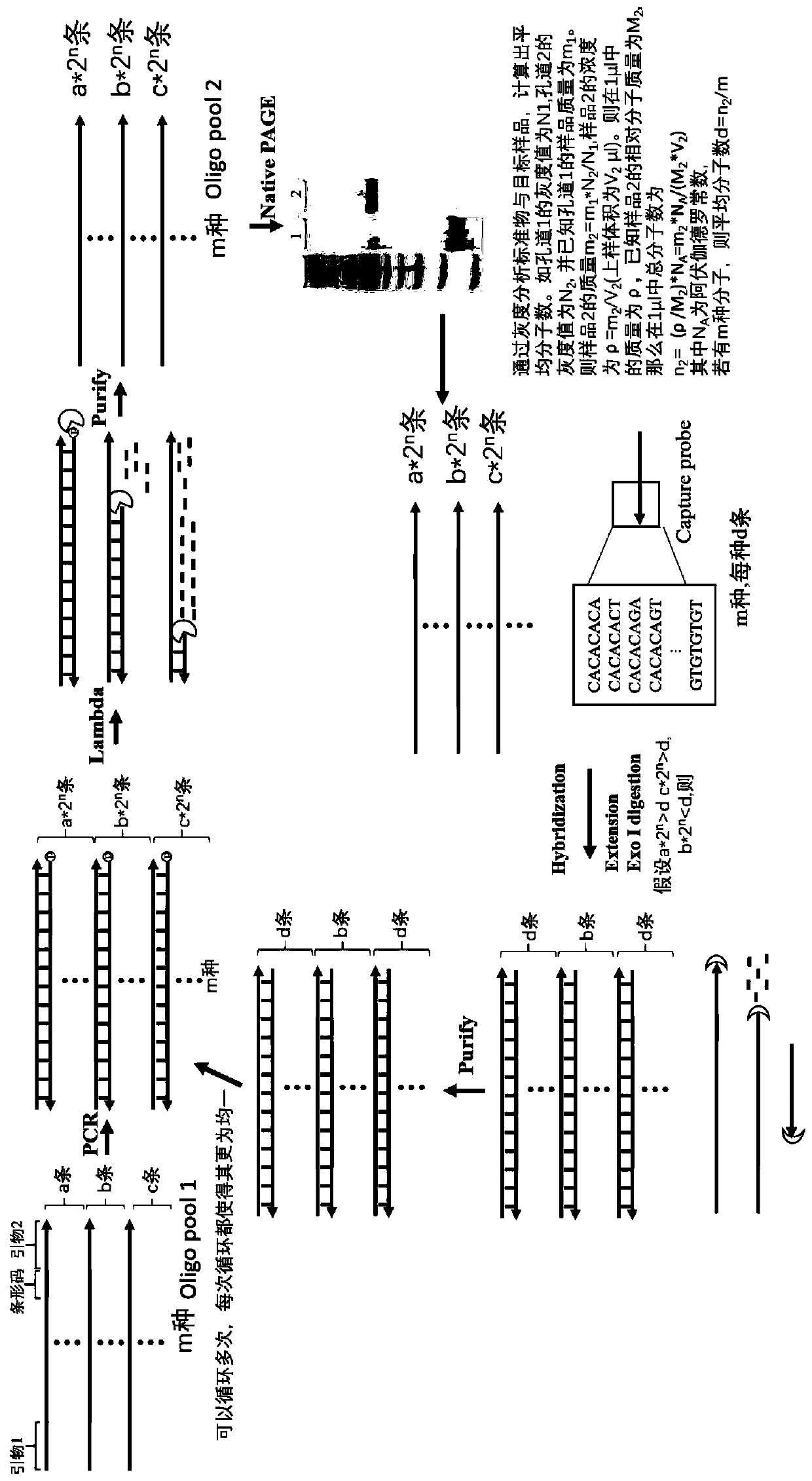
[0082] Embodiment 1: oligo pool is carried out PCR amplification to increase the total number of molecules of oligo pool
[0083] ①PCR system and procedures
[0084] table 3
[0085] Component 50μL Final Conc. h 2 o
21.5 5хQ5 Reaction buffer 10 1х dNTPs (2.5mM) 4 0.2mM F1 (100μM) 2 4μM R1 (100μM) 2 4μM Oligo pool (0.044ng / μL) 10 0.44ng / 50μL Q5 0.5
[0086] F1 is a sequence in which the 1-5 bases at the 5' end are modified with sulfur on the basis of the sequence shown in SEQ ID NO:3, such as C*T*A*CTCCCACTCGTCTATCT; (Lambda exonuclease is 5' non-phosphorylated The modified oligonucleotide has a particularly weak cutting ability. In order to prevent its degradation, the present invention preferably performs sulfur modification on the base at the 5' end of the sequence of F1, * indicates the modified base);
[0087] R1 is the sequence for modifying the phosphate group at the 5' end on the basis of the...
Embodiment 2
[0092] Example 2: Lambda Exonuclease degrades back to single-stranded DNA oligo pool
[0093] (1) Reaction system
[0094] Table 4
[0095] Component 30μL Final Conc. h 2 o
15.5 - 10хLambda exonuclease reaction buffer 3 1х PCR product(above)(69ng / μL) 10 23ng / μL Lambda Exonuclease (5U / μL) 1.5 0.25U / μL
[0096] After incubating at 37° C. for 3 h, EDTA was added to make the final concentration to 10 mM to terminate the reaction.
[0097] (2) Purify and recover the product through column purification according to the operation steps of the instructions of Eastep Gel and PCR Cleanup Kit.
[0098] (3) Dissolve in 40 μL DNase / RNase-free H2O.
[0099] (4) 12% polyacrylamide gel electrophoresis to verify ssDNA (stained with SYBR Gold for 20 minutes), and at the same time, add a known amount of standard DNA and measure the concentration of ssDNA by grayscale analysis. The results of polyacrylamide gel electrophoresis are shown in ...
Embodiment 3
[0102]Example 3: Capture probe captures ssDNA
[0103] The number of molecules of the corresponding capture probe (complementary to the sequence of the primer 2 region and the specific barcode region) is calculated as the average number of molecules in Example 2 (the number of molecules of the capture probe is converted from a certain concentration provided by the manufacturer, N=c*v*N A , c is the concentration of the capture probe, v is the volume to be added to the capture probe) to capture ssDNA, the ssDNA above the average number of molecules is not captured and will be in a free state, while the ssDNA below the capture probe will be all captured, Excess corresponding capture probes are free. After the capture is completed, polymerase polymerase, and then the free ssDNA and capture probes are degraded by exonuclease I. In this way, the number of DNA molecules of various types in the library tends to the average number, and the concentration of various types of DNA is sh...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com