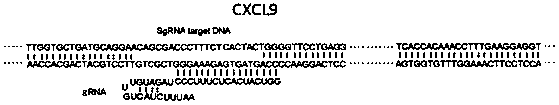Method for screening biallelic mutant cell lines by Cas12a protein
A cell line and allelic technology, applied in the medical and biological fields, can solve the problems of inability to distinguish monoallelic mutations and biallelic knockout strains, time-consuming, labor-intensive, time- and money-increasing costs, etc. The effect of reducing the time and money cost of screening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0024] In order to express the present invention more clearly, the present invention will be further described below in conjunction with the accompanying drawings.
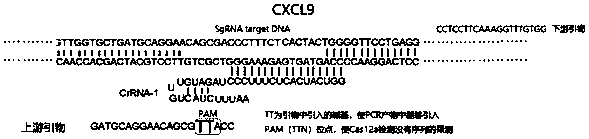
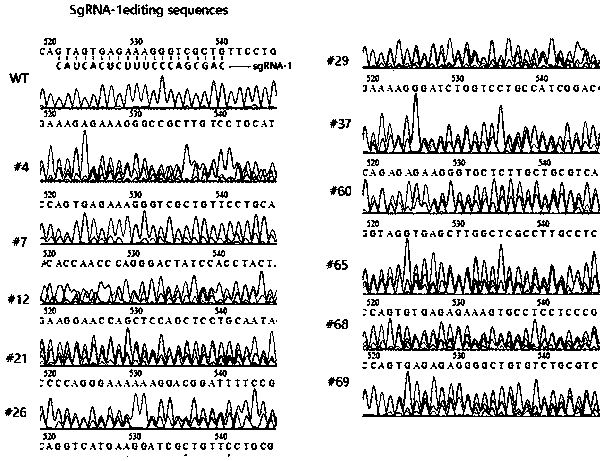
[0025] see figure 1 , the Cas12a / Cpf1 protein used in this application also belongs to the CRISPR-Cas family and has an endonuclease function similar to Cas9. At the same time, the combination of Cas12a-crRNA complex with complementary single-stranded DNA or double-stranded DNA activates its powerful Non-specific ssDNA trans-cleavage activity, labeling ssDNA with a fluorescence quenching (FQ) reporter group can be developed into a nucleic acid detection tool. When Cas12a-crRNA matches the nucleic acid to be detected, Cas12a cuts ssDNA-fluorescent quenching reporter group If the Cas12a-crRNA does not match or does not completely match the nucleic acid to be detected, Cas12a does not cut or partially cuts the ssDNA-fluorescence quenching reporter group, and there is no fluorescence or weak fluorescence.
[0026] se...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com