Disease and metabolite network construction method based on similarity calculation
A technology of similarity calculation and network construction, applied in biological systems, bioinformatics, informatics, etc., can solve problems such as difficult to analyze disease mechanisms, and achieve the effect of eliminating low efficiency and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
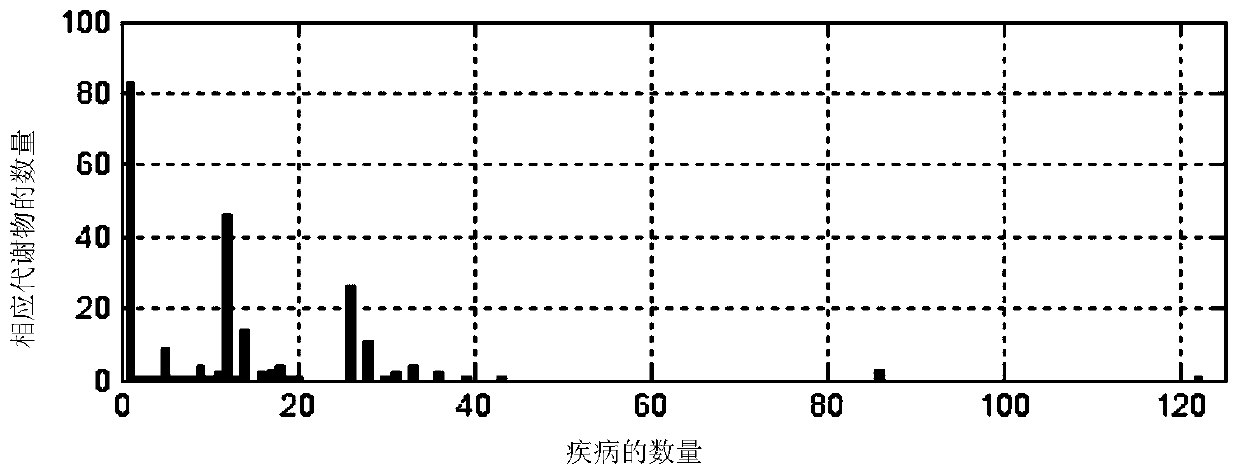
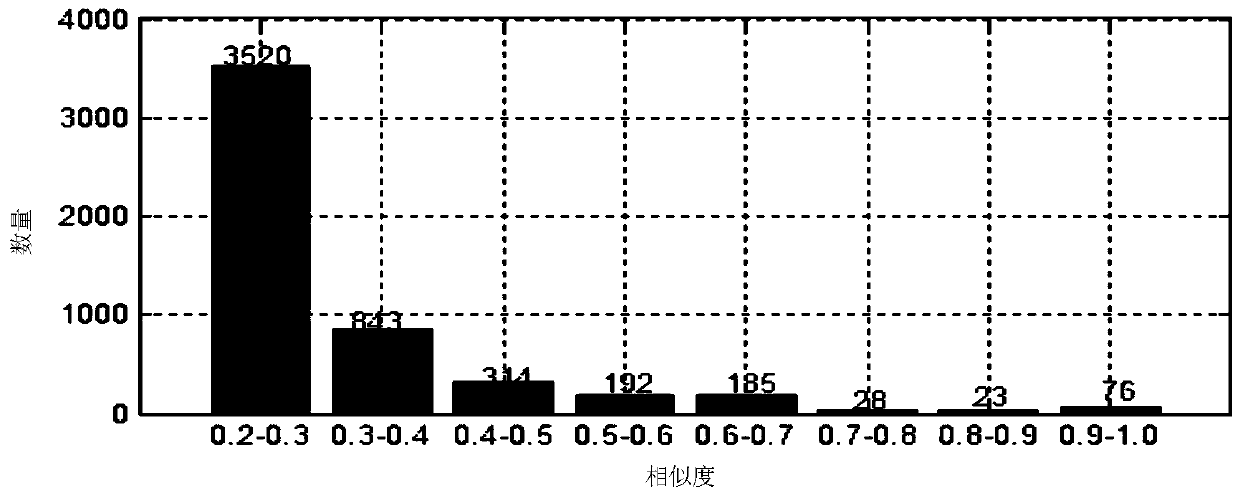
Image
Examples
Embodiment 1
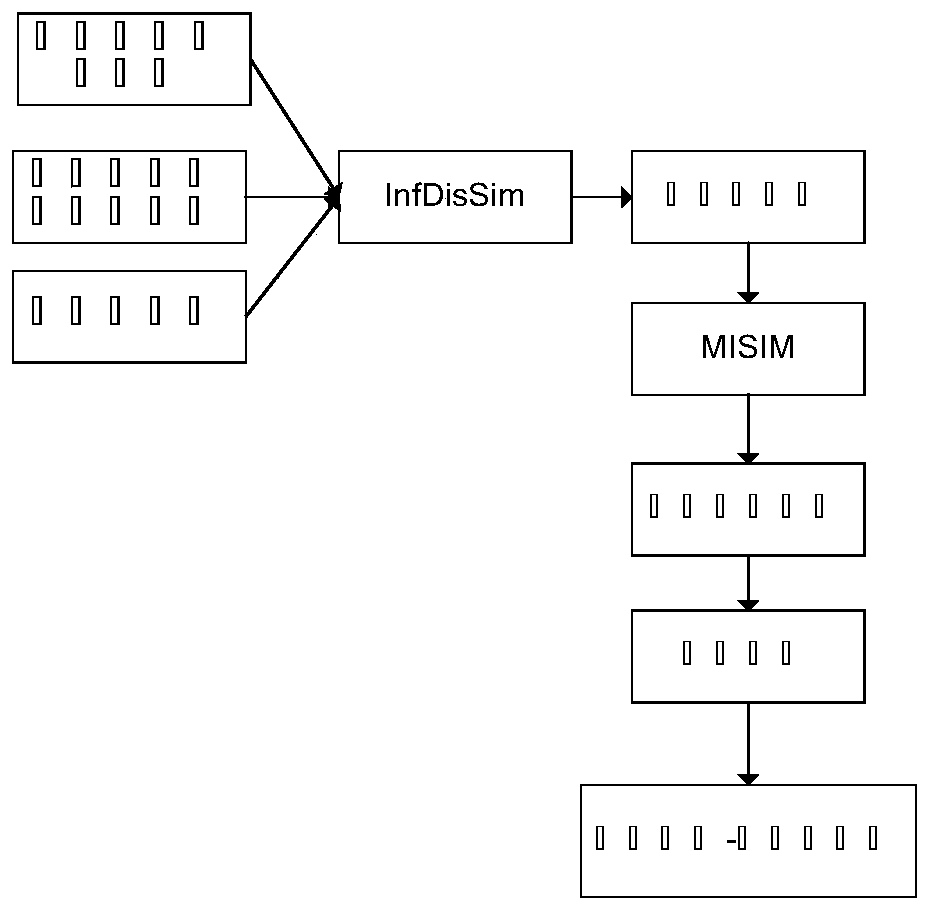
[0041] Embodiment one: if figure 1 As shown, the method for constructing a disease and metabolite network based on similarity calculation involved in this embodiment, the specific steps are:
[0042] Step 1: Obtain information on different diseases and metabolites;
[0043] Step 2: Obtain known metabolites associated with the disease;
[0044] Step 3: Use the InfD-isSim method to find out the similarity of different diseases;
[0045] Step 4: Introduce the MISM method to obtain the similarity of metabolites;
[0046] Step 5: Establish a network of metabolite similarity;
[0047] Step 6: Discover new disease-metabolite relationships by random walk.
Embodiment 2
[0048] Embodiment two: if figure 1 As shown, in the method for constructing a disease and metabolite network based on similarity calculations involved in this embodiment, the acquisition of steps 1 and 2 uses three data sets, which are the human metabolome database, disease ontology and national Biomedical Ontology Center.
[0049] To obtain basic relationships between metabolites and diseases, we use the following three datasets: Human Metabolome Database, National Center for Biomedical Ontology, and Disease Ontology.
[0050] Human Metabolome Database
[0051] We downloaded metabolite data from the Human Metabolome Database (HMDB). The latest and most complete dataset involving more than 40,000 metabolomes. It contains three kinds of data information: chemical data, clinical data and biochemical data. They gathered this information from thousands of public sources.
[0052] The data we get are disease-related metabolites, and they have many complex files. Therefore, we...
Embodiment 3
[0060] Embodiment three: as figure 1 As shown, the method for constructing a disease and metabolite network based on similarity calculation involved in this embodiment, the step three is specifically:
[0061] Each disease has several relevant metabolites, if the number of metabolites is N, the weight vector t1 will be:
[0062]
[0063] is the weight vector of t1, w 1,i is the weight fraction of t1 in the i-th dimension.
[0064] Use cosine to calculate the similarity of diseases, as follows:
[0065]
[0066] Finally, the disease similarity is defined as:
[0067]
[0068] G 1 G 2 Metabolites of t1 and t2 are represented.
[0069] There is some similarity between diseases, and the similarity often arises from molecular origins. The interaction of protein-coding genes can reflect the mechanism of disease to some extent. Therefore, the similarity of diseases can be calculated by the genes behind the diseases.
[0070] To obtain the similarity of diseases, w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com