Method and device for repairing genome sequencing and assembling results, and storage medium
A genome sequencing and genome assembly technology, which is applied in the field of genome sequencing and assembly result repair, and can solve problems such as no correction, incomplete assembly sequence, and loss of assembly sequence.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
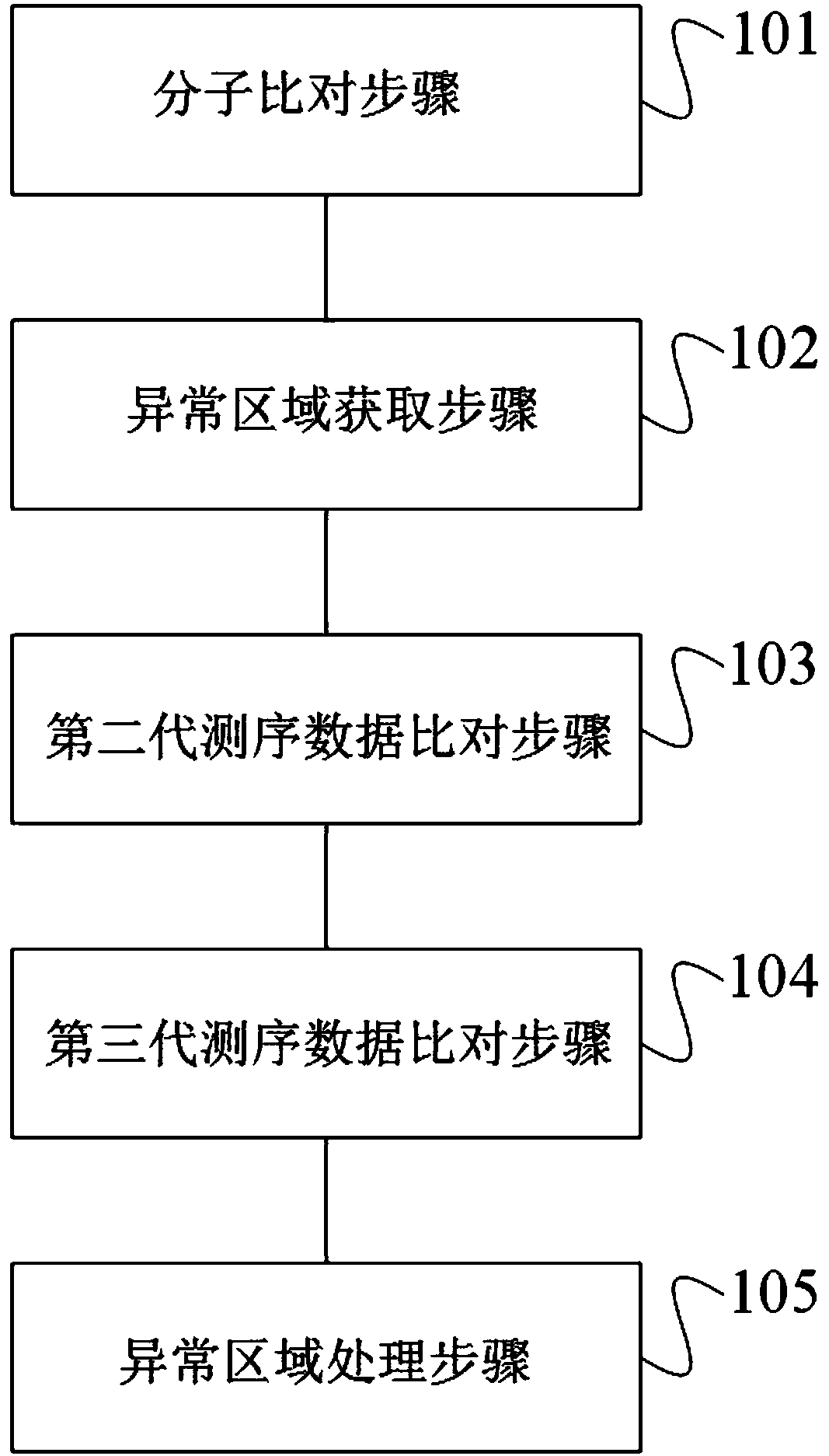
[0073] In this example, for a cereal plant genome with a size of about 2.3Gb, during the genome assembly process, when using the data of the second-generation insert fragment library with a size of 450bp and 800bp to use Pilon software to correct errors and fill holes in the genome, it was found that there were 8 Gap sequences are filled, and the length of these gap sequences is greater than 3k, and even some regions with a gap length of more than 40k are also filled. In order to verify the reliability of these filled sequences, perform the following processing according to the genome sequencing assembly result repair method of this application:
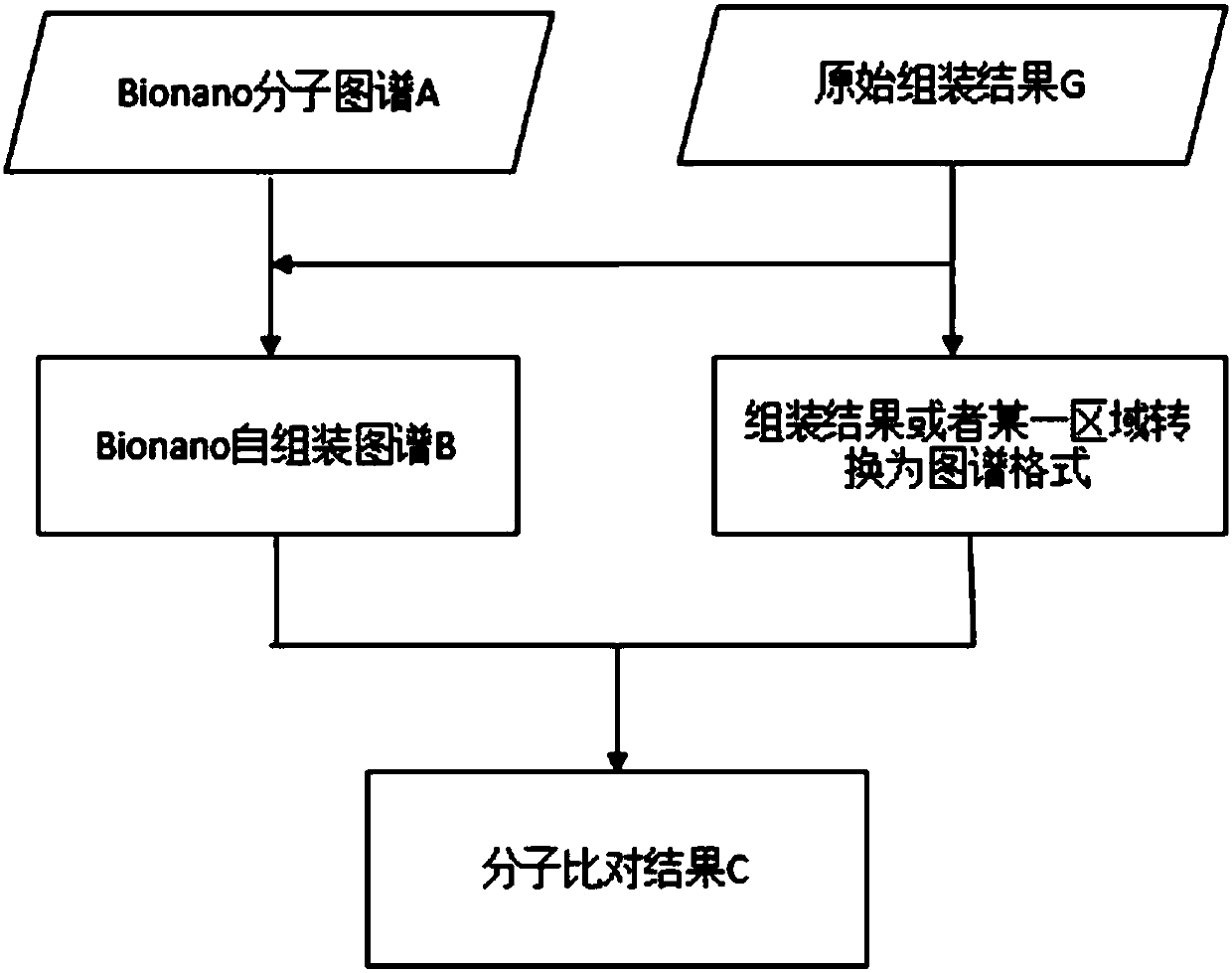
[0074] (1) Molecular comparison step
[0075] 1) Reference sequence preparation: convert the sequence file of the genome assembly result into a file consisting of the position of the corresponding restriction site; specifically, mark the filled sequence ID information, and the position coordinates of the corresponding filled region, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com