Tau modulators and methods and compositions for delivery thereof
A modulator, tau protein technology, applied in drug combinations, peptide/protein components, chemical instruments and methods, etc., can solve problems such as promoting toxicity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0187] Example 1: MAPT repressor
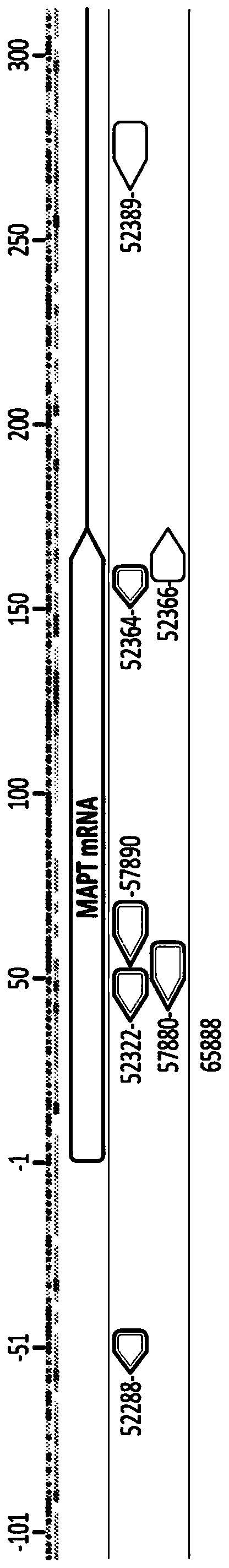
[0188]About 185 zinc finger proteins engineered substantially as described in U.S. Patent No. 6,534,261; U.S. Patent Publication Nos. 2015 / 0082093; 2013 / 0253040; and 2015 / 0335708 were screened and ZFPs bound to their MAPT targets location. The zinc finger proteins 52288, 52322, 52364, 52366, 52389, 57880, 57890 and 65888 targeting mouse MAPT (see Tables 1 to 3 below) were selected for further study. The phosphocontact mutants listed for 65888 are as previously described (see eg US Patent Application No. 15 / 685,580). Table 1 shows the recognition helices of the DNA binding domains of these ZFPs, and the target sequences of these ZFPs. A panel of ZFPs was also made to target MAPT sequences shared between mouse and human genes. These are shown in Table 2. Table 3 shows parental and derived ZFPTFs in which the ZFP backbone has been mutated at the indicated positions to remove potential non-specific phosphocontacts. ZFPs were assessed by stan...
Embodiment 2
[0200] Example 2: Tau repression in mouse neurons
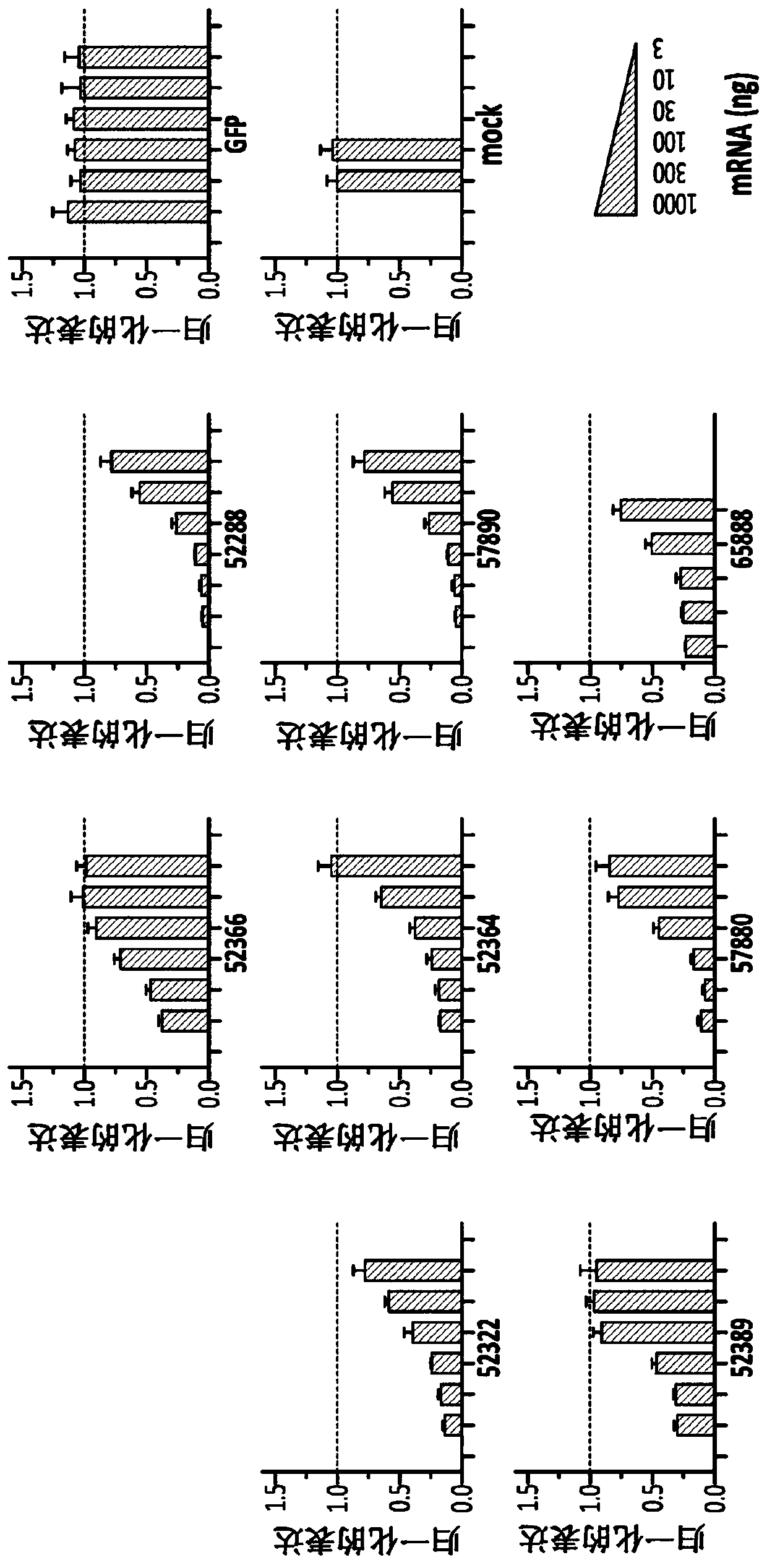
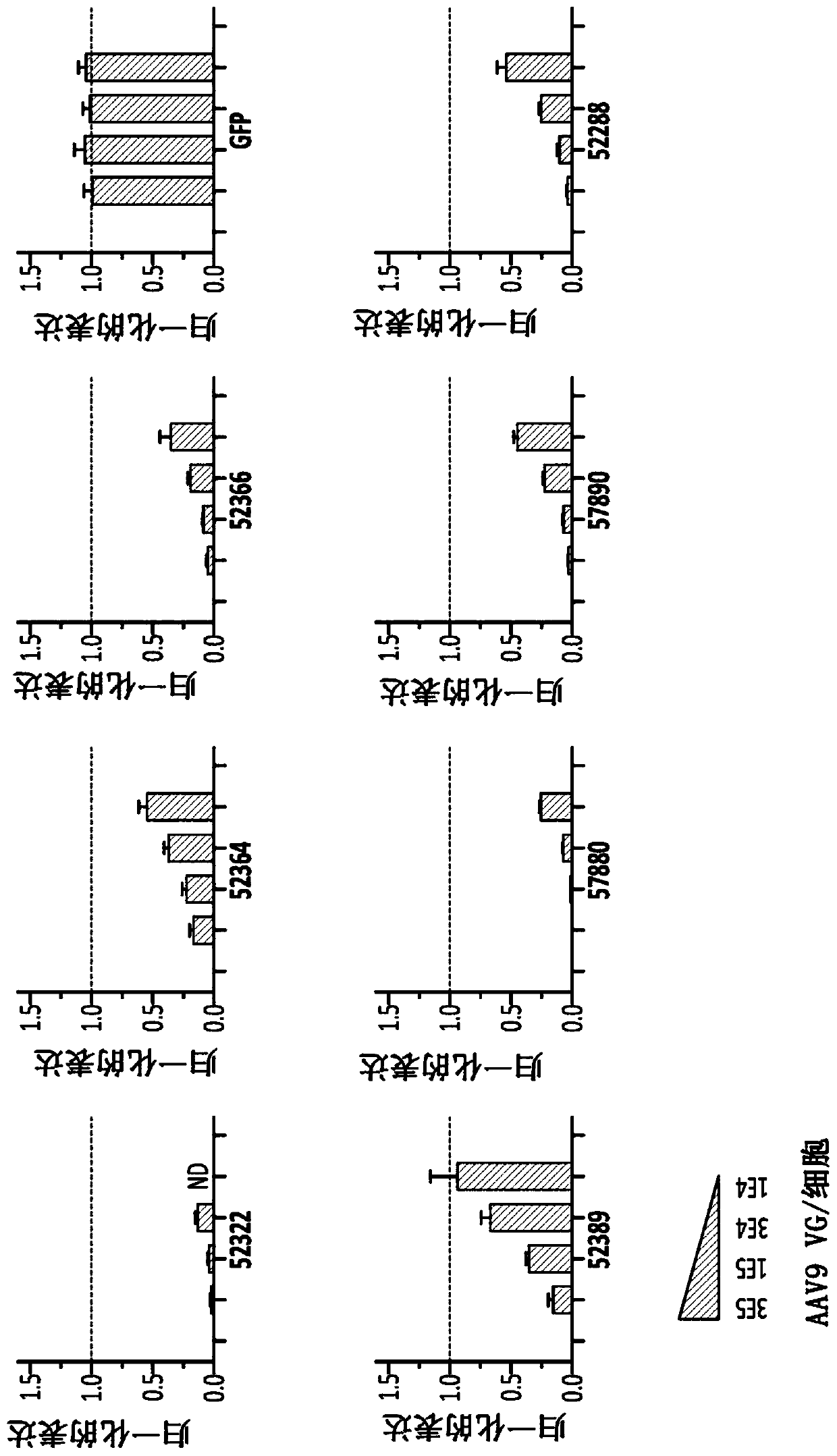
[0201] All ZFP repressors were cloned into rAAV2 / 9 vectors using the CMV promoter to drive expression. Virus was produced in HEK293T cells, purified using a CsCl density gradient, and titrated by real-time qPCR according to methods known in the art. Primary mouse cortical neurons in culture were infected with purified virus at 3E5, 1E5, 3E4 and 1E4VG / cell. After 7 days, total RNA was extracted and the expression of MAPT and two reference genes (ATP5b, EIF4a2) were monitored using real-time RT-qPCR.
[0202] like Figure 1C As shown in , all ZFP-TFs encoding AAV vectors were found to efficiently repress mouse MAPT over a broad infectious dose range, with some ZFPs reducing targets by >95% at multiple doses. In contrast, no MAPT repression was observed for rAAV2 / 9CMV-GFP virus or mock-treated neurons tested at equivalent doses (see Figure 1).
[0203] Mouse cortical neurons were infected with 3E5, 1E5 and 3E4 VG / cell using ...
Embodiment 3
[0205] Example 3: Specificity of MAPT repression
[0206] The overall specificity of the ZFP-TFs shown in Tables 1 and 2 was assessed in mouse Neuro2A cells by microarray analysis. Briefly, 300 ng of ZFP-TFs encoding mRNA were transfected into 150,000 Neuro2A cells in biological quadruplicate. After 24 hours, total RNA was extracted and processed according to the manufacturer's protocol (Affymetrix Genechip MTA1.0). Raw signals from each probe set were normalized using robust multiarray averaging (RMA). Analysis was performed using Transcriptome Analysis Console 3.0 (Affymetrix) with the "gene level differential expression analysis" option. ZFP-transfected samples were compared to samples that had been treated with an irrelevant ZFP-TF (which did not bind to MAPT target sites). Transcript (probe set) change information (change call).
[0207] like image 3 As shown in , SBS#52322 repressed 5 genes except MAPT and caused an increase of 3 other genes. SBS#52364 and #52366...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com