Cluster analysis method based on peripheral blood plasma free DNA nucleosome footprint difference, and application thereof
A cluster analysis and nucleosome technology, applied in the biological field, can solve problems such as treatment failure, patient suffering, and inability to alleviate the condition, and achieve high coverage and good accuracy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Example 1: Cluster analysis of chemotherapy-sensitive group and chemotherapy-insensitive group of lung cancer
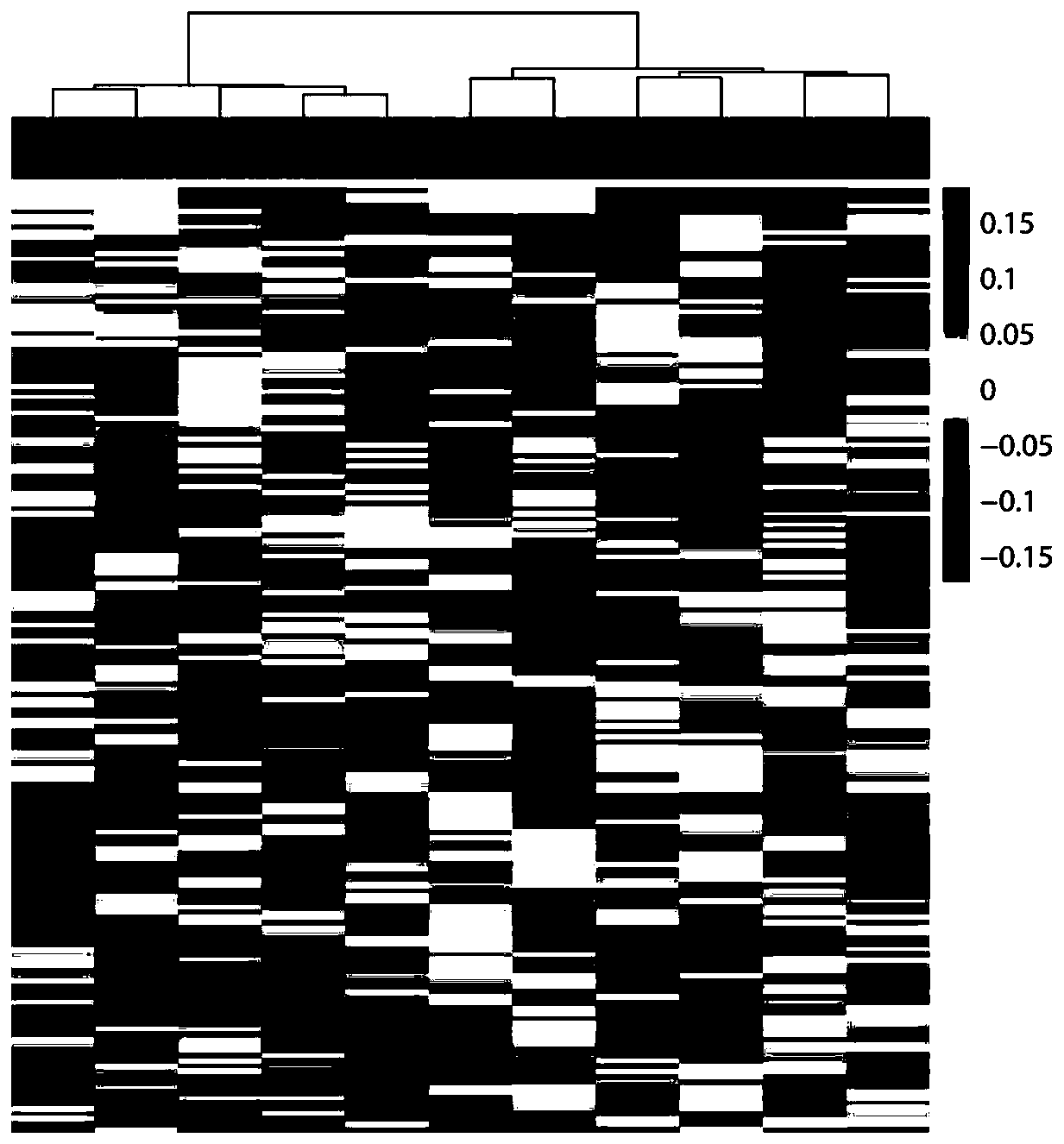
[0025] Based on the high-throughput sequencing data of cell-free DNA in peripheral blood plasma of 5 lung cancer chemotherapy-sensitive and 6 insensitive patients, the coverage difference analysis of TSSs and TTSs regions was performed, and it was found that chemotherapy-sensitive patients had different TSSs and TTSs from insensitive patients There were 178 regions (see Table 2), of which the coverage of 88 differential TSSs regions was upregulated in the sensitive patient group, and the coverage of 90 differential TSSs regions was downregulated in sensitive patients. By using differentially expressed genes for unsupervised hierarchical clustering analysis, it was found that based on 178 differential genes, samples can be clustered into sensitive and insensitive categories, and the patterns of coverage differential genes of the two groups are significantly diff...
Embodiment 2
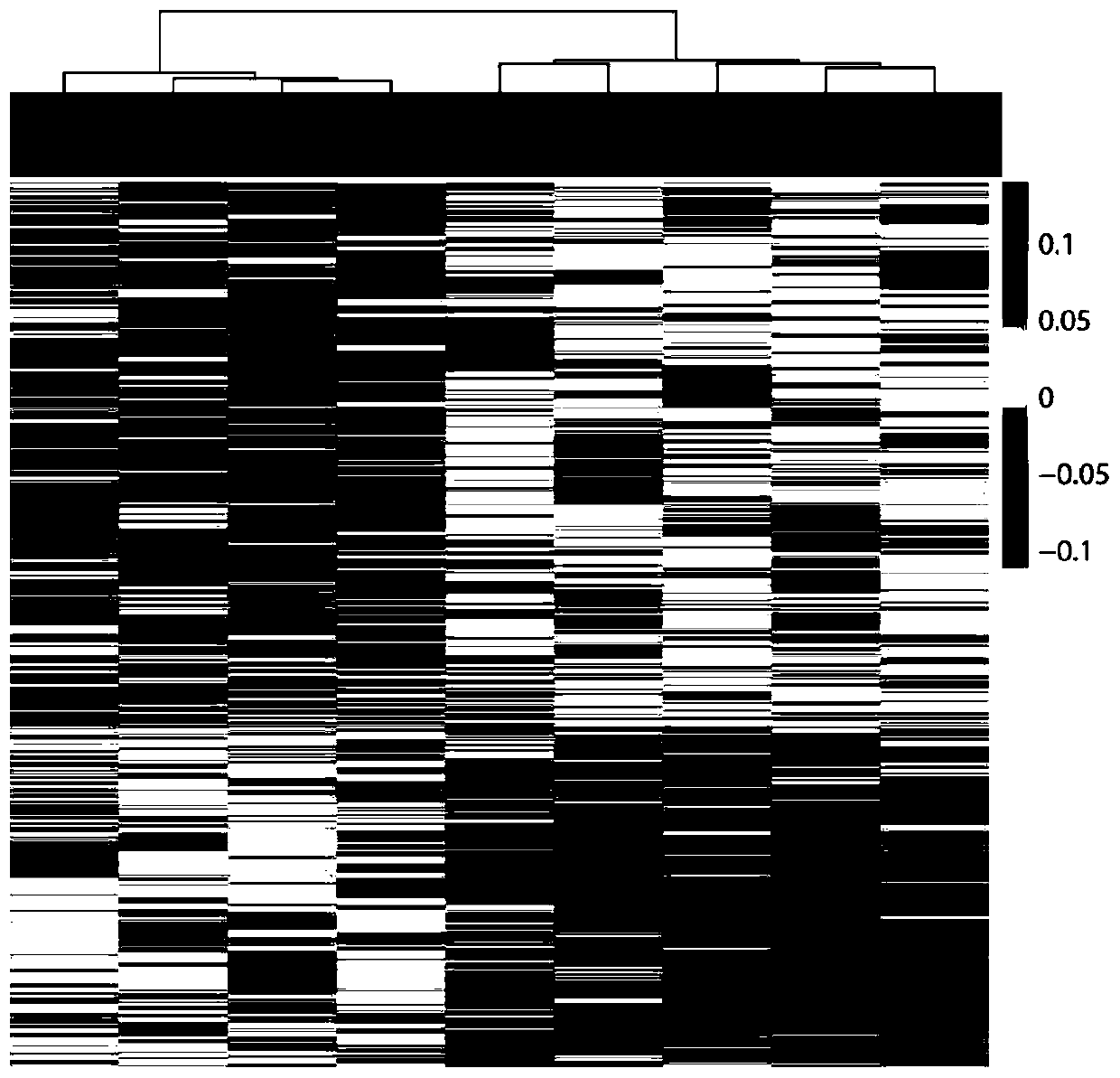
[0042] Example 2: Cluster comparative analysis based on the complete remission group and the insensitive group of neoadjuvant chemoradiotherapy for colorectal cancer DNA high-throughput sequencing data (see Table 3), and the coverage difference analysis of TSSs and TTSs regions, found that there were 597 TSSs and TTSs regions (see Table 4) that were different between radiotherapy and chemotherapy-sensitive patients and insensitive patients (see Table 4), of which 368 The coverage of regions with 10 differential TSSs was upregulated in the sensitive patient group, and the coverage of 229 regions with differential TSSs was downregulated in sensitive patients. Through unsupervised hierarchical clustering analysis using differentially expressed genes, it was found that based on 597 differential genes, samples could be clustered into two categories: complete remission and insensitivity, and the patterns of coverage differential genes of the two groups were significantly different. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com